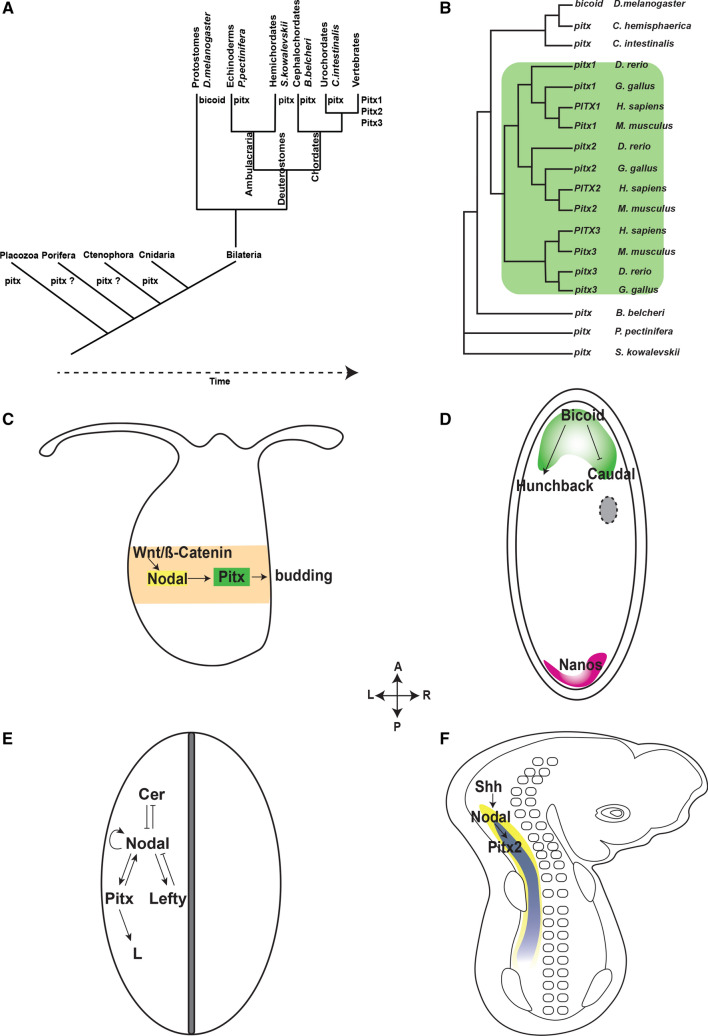
Fig. 1.
The evolutionarily conserved Pitx gene family. a Pitx genes have been identified in most animal groups from the simplest to the most complex in the animal kingdom. Among the four basal phyla, pitx genes have not been determined in Porifera and Ctenophora. In Placozoa, pitx expression has been evidenced but no associated function has been reported. Pitx genes in members of the Cnidaria and Bilateria have been well characterized. In Protostomes, Bicoid is the homolog of Pitx. Only vertebrates possess Pitx1, Pitx2, and Pitx3. b Phylogenetic tree of the Pitx gene family in animal by protein sequence alignments. All genes are represented in human (H. sapiens), mouse (M. musculus), chick (G. gallus), zebra fish (D. rerio), fruit fly (D. melanogaster), jellyfish (C. hemisphaerica), sea vase (C. intestinalis), amphioxus (B. belcheri), starfish (P. pectinifera), and acorn worm (S. kowalevskii). There is an incredible conservation of PITX protein sequences of species belong to different groups of animals. There are two main duplication events. The first duplication gives rise to Pitx3 and a precursor of Pitx1/2 and the latter of which ultimately becomes Pitx1 and Pitx2 in the second duplication event. c Pitx in molecular pathway that regulates asymmetrical branching in hydra. The initial broad expression of Wnt/ß-Catenin (orange) induces a more localized expression of Nodal (yellow) around the budding site. Nodal activates the transcription of Pitx (green) that is restricted to budding site to initiate branching. d Anterior segment specification by bicoid in fruit fly (D. melanogaster). Two maternal genes that specify the anterior and posterior of the body plan of fruit fly are bicoid (green) and nanos (pink). Before fertilization, bicoid mRNA is most abundant in the anterior while nanos is highly expressed in the opposite pole of the egg. Upon fertilization, bicoid mRNA is translated to protein which enhances the expression of hunchback and other genes associated with the anterior segment while at the same time inhibit the transcription of genes regulating the posterior region. e Left–right asymmetry in non-vertebrate deuterostomes. The molecular pathway that determines the left-side morphology in invertebrates deuterostomes starts with asymmetric expression of Nodal (yellow) that activates the transcription of Pitx (green). PITX proteins then bind to the enhancer regions and activate downstream target genes that are responsible for regulating left–right asymmetry. f Left–right asymmetry in vertebrates. In chick embryo at stage 8, Pitx2 (blue) is expressed broadly in the left lateral plate mesoderm but is completely absent in the right counterpart. Pitx2 acts downstream of Nodal and Shh