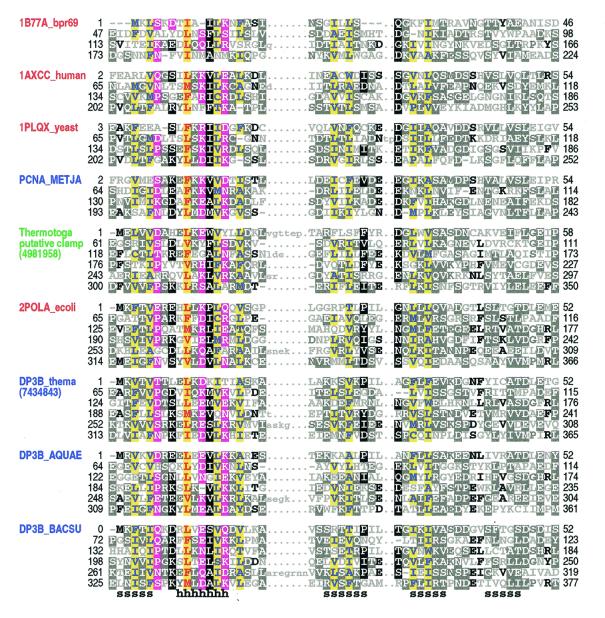
Figure 1.
Representative alignment of known and putative processivity fold proteins. Names of proteins of known structure are shown in red, of previously predicted clamp proteins in blue and of a putative clamp protein from T.maritima in green (see text). Proteins of known structure are designated by their pdb identifiers; other sequences are designated by their SwissProt identifiers or gi numbers. Abbreviations used for organism names are: bpr69, bacteriophage R69; METJA, Methanococcus jannaschii; ecoli, Escherichia coli; thema, Thermotoga maritima; AQUAE, Aquifex aeolicus; BACSU, Bacillus subtilis. The transition probability pairs (expressed in 200th nats) used to obtain the HMM parameters from secondary structure propensities are: match-to-insert, 350–1700; insert-to-insert, 20–150; match-to-delete, 400–500; delete-to-delete, 40–700 (see Materials and Methods). The first repeat in 1B77A_bpr69, which was not found by the sampler, was determined through optimum alignment against a HMM of four repeats. For each aligned column, elevated residues (i.e. with binomial tail probabilities <0.01) and related, marginally conserved residues (with tail probabilities <0.05) are indicated using the following automated hierarchical coloring scheme (see 8). Columns with ≥1.25 bits of information and hydrophobic, red on yellow highlight. Columns with 0.75–1.25 bits of information: hydrophobic, blue on yellow highlight; non-hydrophobic, magenta highlight. Other columns: ≥70% hydrophobic, yellow highlight; >66% conserved, black highlight; 50–66% conserved, dark gray highlight; 33–50% conserved, black; <33% conserved, dark gray; unconserved, light gray. Note that this coloring scheme is based on the full alignment of about 475 repeats. Consensus structural assignments based on known structures are shown below the alignment (h, helix; s, strand).