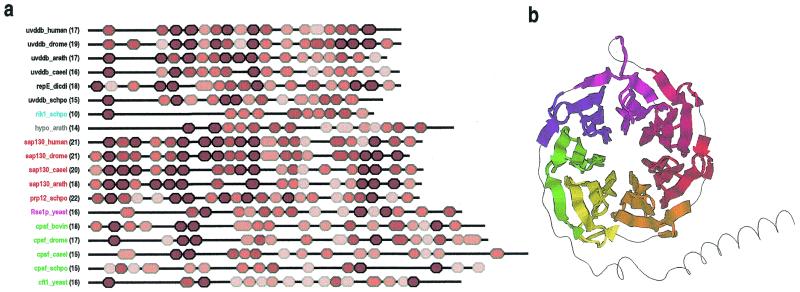
Figure 5.
(a) Domain architectures for UV-DDB-127 repeat proteins. Repeats are colored red proportional to their likelihood scores using lighter shades for less conserved repeats. Protein names are color coded according to families. The number of repeats are indicated in parentheses. GenBank identifiers for the sequences are: uvddb_human, 4503279; uvddb_drome, 4928452; uvddb_arath, 7267302; uvddb_caeel, 7506084 ; repE_dicdi, 2130171; uvddb_schpo, 7492324; rik1_schpo, 7493406; hypo_arath, 6671952; sap130_human, 3540219; sap130_drome, 7292001; sap130_caeel, 7505161; sap130_arath, 7019653; prp12_schpo, 6451681; Rse1p_yeast, 6323592; cpsf_bovin, 1706101; cpsf_drome, 7303176; cpsf_caeel, 7105681; cpsf_schpo, 7492482; cft1_yeast, 6320507. Abbreviations used for organism names are: drome, Drosophila melanogaster; arath, Arabidopsis thaliana; caeel, Caenorhabditis elegans; dicdi, Dictyostelium discoideum; schpo, Schizosaccharomyces pombe. The transition probability pairs (expressed in 200th nats) used to obtain the HMM parameters from secondary structure propensities are: match-to-insert, 150–1300; insert-to-insert, 10–70; match-to-delete, 200–600; delete-to-delete, 150–350 (see Materials and Methods). (b) Structural regions of a WD40 repeat protein (human transducin β-chain, pdb 1GG2B) (55) that align with seven UV-DDB-127 repeats defined by the HMM corresponding to the alignment in Figure 4. A randomly shuffled transducin sequence was appended to the original sequence to increase the likelihood of misalignment. All seven UV-DDB-127 HMM repeats (shown using seven distinct colors in the figure) align with the transducin WD40 repeats in a manner consistent with the proposed structural arrangement (see text).