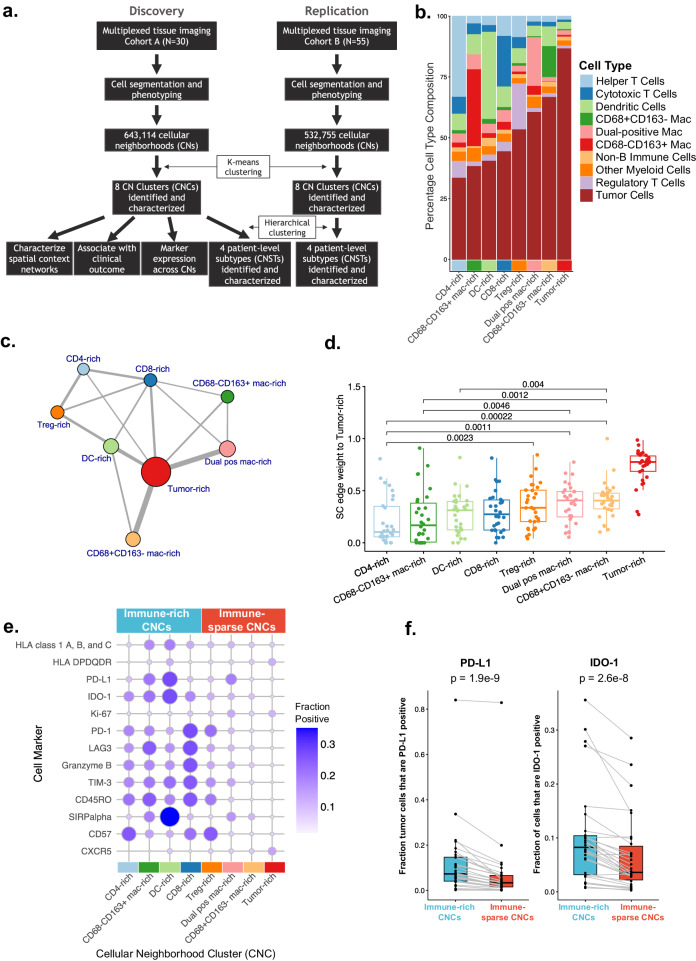
Fig. 1. Study overview and spatial characterization of cellular neighborhood clusters (CNCs).
a Study design and analysis flow. b Cell type composition of each cellular neighborhood cluster (CNC). The CNCs are ordered by increasing tumor content. c Aggregate spatial context networks across all samples in Cohort A. Each CNC is represented as a colored node, with node sizes proportional to the total number of CNCs in the samples. Edges between two CNCs are shown if >10% of CNs within the two CNCs were in close proximity. Edge thickness (weight) is proportional to this percentage of close proximity between pairs of CNCs. Self-edges are not included for visual clarity. d Spatial context network edge weights (fraction of CNs within each CNC in close proximity) between tumor-rich CNs and all other CNs, for each sample in Cohort A. This is a quantified depiction of the edge weights shown in c, between tumor-rich CNs and all other CNs. e Relative expression of functional markers across all eight CNCs, for Cohort A. CNCs are ranked based on immune cell abundance and labeled as immune-rich and immune-sparse. f The expression of PD-L1 or IDO-1 on tumor cells within each sample. p values from Wilcoxon paired test.