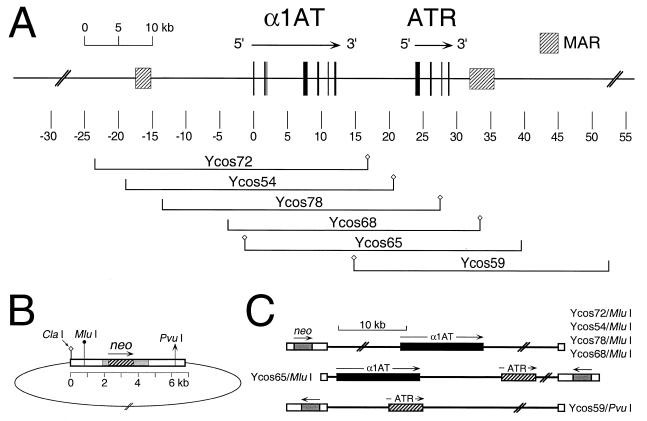
Figure 1.
Genomic map of the α1AT region and cosmids used in this study. (A) Physical map of the α1AT-ATR region. The map is drawn to scale, with position zero defined as an EcoRI site in macrophage-specific exon IA of α1AT (5,35). Exons are indicated by black boxes, and MARs as hatched boxes. The positions and names of relevant cosmids used in this study are shown below the map. Small diamonds on either side of the genomic inserts represent the diagnostic ClaI site of the cosmid vector. (B) Schematic representation of the SuperCos1 vector ligated to a genomic insert. White boxes indicate λ or plasmid sequences. The bacterial aminoglycoside phosphotransferase (neo) gene (hatched box) is under the control of SV40 promoter and polyadenylation signals (gray boxes). Relevant restriction sites used to determine insert orientations and/or to linearize constructs for electroporation are indicated. (C) Diagrammatic representation of various linearized cosmid clones, showing the presumptive genomic organization of single-copy, intact integrants. The relative positions and orientations of neo (gray boxes) and α1AT (black boxes) and/or ATR (hatched boxes) in the various linearized templates are indicated.