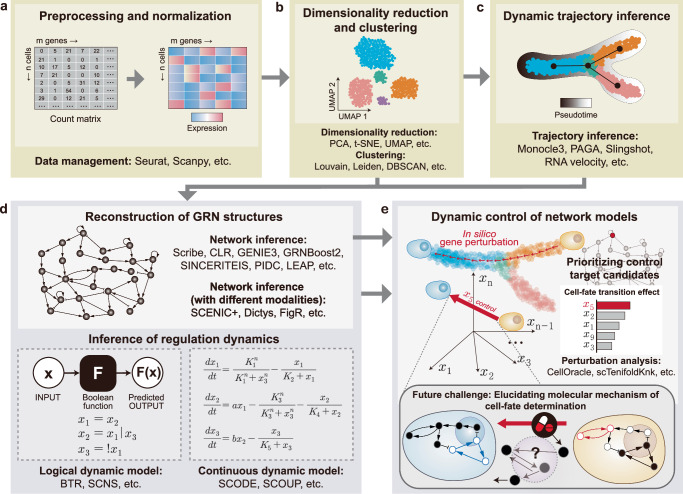
Fig. 2. A process for constructing mathematical models using scRNA-seq data.
a Utilization of scRNA-seq data in studying cell-fate determination. Initially, transcriptomic data is initially preprocessed to build a count matrix of cells by genes derived from raw data processing steps. Next, the count matrix is preprocessed using tools such as Seurat and Scanpy. b Identification of specific cell fates. Dimensionality reduction methods are applied to handle the complexity of the data, and clustering algorithms group cells with shared identities. Subsequently, specific cellular states related to the cell fates of interest are identified. c Inference of dynamic trajectories to reveal cell-fate changes. Cellular trajectories are derived from gene expression changes. In particular, Monocle3 and RNA velocity are generally used to infer cellular dynamic trajectories from scRNA-seq datasets. d Inference of GRN structures and regulation dynamics. Molecular interactions are obtained from the processed data. Furthermore, a network model can be constructed by inferring the critical regulation dynamics involved in cell-fate determination by discrete or continuous mathematical formalisms. e Dynamic control of network models. In silico gene perturbation analysis with tools like CellOracle and scTenifoldKnk can predict the effects of control target candidates. However, they lack insight into regulatory dynamics represented by attractor landscapes. To tackle this limitation, the future direction of research should progress beyond applying control theory to elucidate the molecular mechanisms underlying cell-fate changes.