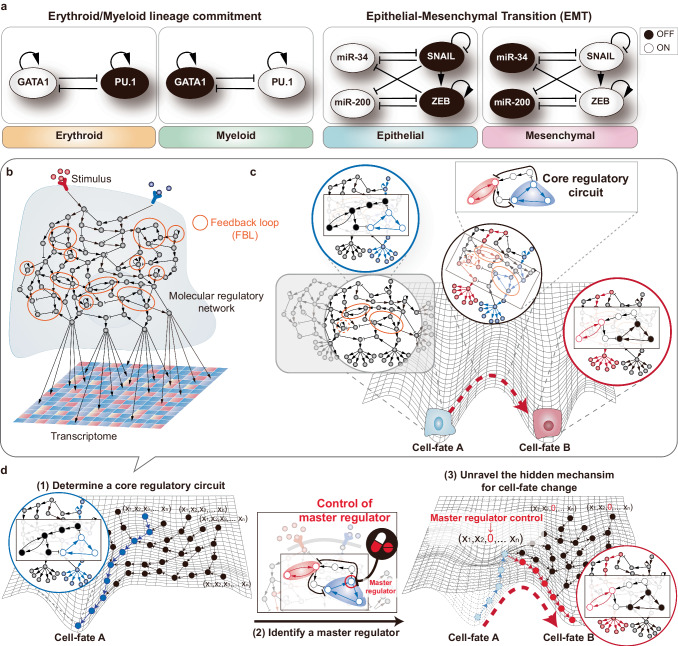
Fig. 3. Core regulatory circuits in a dynamic network model.
a Examples of key regulatory components for cell-fate determination. b Representation of regulation dynamics at the network-level. The molecular regulatory network is useful to describe the regulation dynamics inferred from transcriptome data in response to a stimulus. From these molecular regulations, all feedback loops are investigated to find out an essential subnetwork. c Core regulatory circuit for cell-fate determination. As each cell fate is defined by the gene activities within the molecular regulatory network, alteration in a few critical molecular interactions can lead to a shift towards a specific cell fate. In other words, a specific subnetwork composed of interconnected positive feedback loops (referred to as a ‘core regulatory circuit’) is considered the primary driver of cell-fate changes. For example, it could be a toggle switch, a well-known designed circuit in cell-fate decision-making. d Elucidating the molecular mechanism through a core regulatory circuit and its master regulator. Each cell fate can be regarded as an attractor within the attractor landscape that includes the transition trajectory. First, based on detailed analysis as outlined in b, c, a core regulatory circuit essential for cell-fate determination can be extracted. Next, a master regulator that governs the change of cellular states described by the core regulatory circuit can be identified. Finally, the hidden mechanism underlying the transition when the master regulator is controlled can be unraveled.