Figure 2.
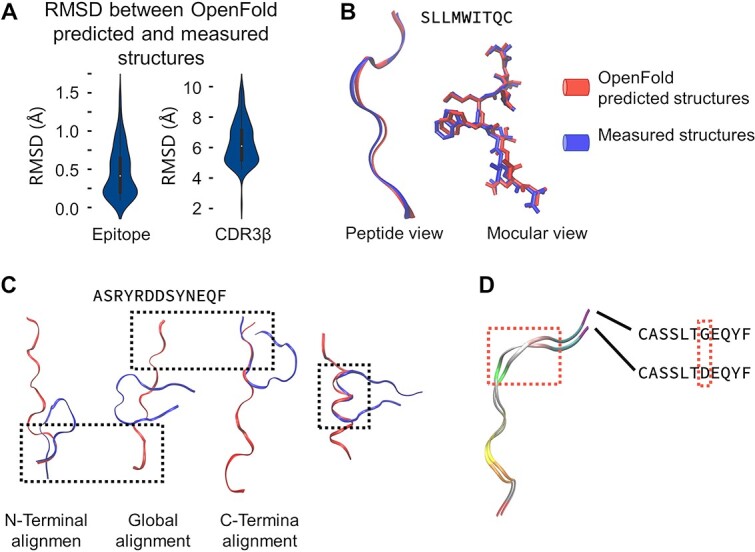
OpenFold predictions of CDR3-β and epitope structures. (A) The RMSD between OpenFold’s predicted structures for epitopes and CDR3-β and the corresponding structures obtained from the RCSB PDB. (B) OpenFold prediction results for epitopes, with the left subfigure depicting the predicted amino acid chain and the right subfigure illustrating the predicted molecular conformation. (C) OpenFold predictions for the CDR3-β structures. OpenFold faces challenges in accurately predicting the conformation of the CDR3-β central segment due to the absence of certain contextual clues that guide protein folding; however, OpenFold has good predictive accuracy for various subsegments. (D) OpenFold’s predictions for sequences with high similarity (differing by a single amino acid) show its ability to discern subtle structural differences. RMSD: root mean square deviation.
