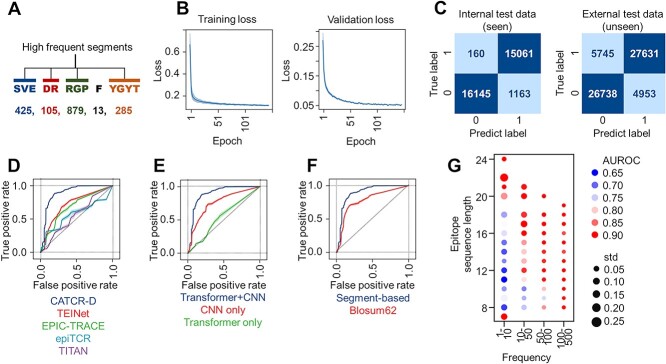
Figure 3.
CATCR-D Prediction of Epitope-CDR3-β Binding. (A) The segment-based coding method, where high-frequency amino acid segments are coded as single characters. (B) Training and validation set losses. (C) Confusion matrix visualizing classification results for the internal and external test sets. (D) Comparison of the AUROC of CATCR-D on the external test set, which includes unseen epitopes and CDR3 sequences, against those of the benchmark methods (TEINet, EPIC-TRACE, epiTCR and TITAN). (E) External test set AUROC comparison between CATCR-D and methods using only a CNN for RCM feature extraction and only a transformer for sequence feature extraction. (F) Performance of segment-based coding versus BLOSUM62 in terms of AUROC on the external test set. (G) AUROC of CATCR-D on the external test set across epitopes of varying lengths and frequencies of occurrence. AUROC: area under the receiver operating characteristic curve.