Abstract
Swine are regarded as “intermediate hosts” or “mixing vessels” of influenza viruses, capable of generating strains with pandemic potential. From 2020 to 2021, we conducted surveillance on swine H1N2 influenza (swH1N2) viruses in swine farms located in Guangdong, Yunnan, and Guizhou provinces in southern China, as well as Henan and Shandong provinces in northern China. We systematically analyzed the evolution and pathogenicity of swH1N2 isolates, and characterized their replication and transmission abilities. The isolated viruses are quadruple reassortant H1N2 viruses containing genes from pdm/09 H1N1 (PB2, PB1, PA and NP genes), triple-reassortant swine (NS gene), Eurasian Avian-like (HA and M genes), and recent human H3N2 (NA gene) lineages. The NA, PB2, and NP of SW/188/20 and SW/198/20 show high gene similarities to A/Guangdong/Yue Fang277/2017 (H3N2). The HA gene of swH1N2 exhibits a high evolutionary rate. The five swH1N2 isolates replicate efficiently in human, canine, and swine cells, as well as in the turbinate, trachea, and lungs of mice. A/swine/Shandong/198/2020 strain efficiently replicates in the respiratory tract of pigs and effectively transmitted among them. Collectively, these current swH1N2 viruses possess zoonotic potential, highlighting the need for strengthened surveillance of swH1N2 viruses.
Keywords: Swine influenza virus (SIV), H1N2, Evolution, Replication, Pathogenicity, Zoonotic potential
Highlights
-
•
Five quadruple reassortant swH1N2 viruses were identified which have close genetic relationship with current human virus.
-
•
The swH1N2 viruses replicates effectively in human, canine, and swine cells and mice.
-
•
A/swine/Shandong/198/2020 replicates efficiently in the respiratory tract of pigs and spreads effectively.
-
•
The swH1N2 viruses pose zoonotic potential, highlighting the need for strengthened surveillance of swH1N2 viruses.
1. Introduction
Swine are known as “mixing vessels” for influenza viruses. In pigs, avian, swine and human influenza viruses can undergo internal genetic recombination, creating new viruses with the potential to infect humans and cause a pandemic, thereby posing a significant threat to public health. Although the evolution speed of the swine influenza virus is not as rapid as that of the human influenza virus (de Jong et al., 2007), the frequent close contact between humans and pigs facilitates the transmission of influenza viruses between the two species.
Studies have demonstrated that the North American triple reassortant swine influenza A virus H1 isolates have the ability to replicate and transmit in ferrets (Barman et al., 2012), making them the standard animal model for assessing the pathogenesis and transmission risk of influenza virus (Belser et al., 2016). This highlights the need for close monitoring of mutant viruses. In addition to mutation and reassortment, influenza viruses can also undergo a relatively rare evolutionary process called homologous recombination. During the replication of the influenza A virus (IAV), genomic RNA and ribonucleoprotein are rapidly packaged to prevent template conversion (Shao et al., 2017). Consequently, homologous recombination is infrequent in influenza viruses (Chare et al., 2003). However, some studies have raised the possibility of controversial homologous recombination during IAV replication. For instance, previous research demonstrated that PB2 can undergo homologous recombination with PA, HA (between human H1N1 subtype strains), and NP (between human H1N1 subtype strains and H3N2 subtype strains) (He et al., 2008). This finding serves as an early warning for the emergence of new pandemic viruses.
The H1N2 virus was initially detected in Japanese swine herds in 1978, and it is believed to have originated from reassortant between human H1N1 and H3N2 viruses (Brown et al., 1998). Since then, this type of recombinant virus has consistently been isolated from pigs (Nerome et al., 1985; Ito et al., 1998; Ouchi et al., 1996). In China, the first reported outbreak of H1N2 swine influenza virus (SIV) occurred in Zhejiang Province in 2004 and led to subsequent swine outbreaks in 2006 (Pulit-Penaloza et al., 2019). Additionally, between 2011 and 2012, a novel triple recombinant H1N2 influenza virus was isolated from pigs in southern China and South Korea. This particular strain contained six internal genes that were derived from the 2009 H1N1 pandemic virus. The recombinant H1N2 influenza viruses continue to be a subject of frequent monitoring and concern. Due to the distinct evolutionary and epidemiological characteristics of human and swine influenza viruses, humans generally lack immunity to endemic viruses that have evolved in pigs. In 2021, a novel recombinant virus was isolated from a one-year-old girl in Taiwan who exhibited fever and other symptoms. This virus harbored fragments of the HA and NA genes that were derived from the swine influenza A (H1N2) virus (Yang et al., 2022).
As the world's largest producer and consumer of pork, China frequently imports pigs from other continents (Zhu et al., 2013; Kong et al., 2014). Consequently, China has a highly complex swine influenza virus ecosystem. To understand the genetic evolution of the current swH1N2 virus in China and assess its zoonotic and pandemic potential, this study has three primary objectives: (i) to investigate the phylogenetic evolution and molecular characteristics of H1N2 swine influenza viruses (swIAVs); (ii) to examine the replication ability of H1N2 swIAVs in vitro; and (iii) to evaluate the pathogenesis and transmission of H1N2 swIAVs in mice and pigs.
2. Materials and methods
2.1. Sample collection and virus isolation
From October 2020 to April 2021, we conducted surveillance on swine influenza viruses in swine farms located in Guangdong, Yunnan, and Guizhou provinces in southern China, as well as Henan and Shandong provinces in northern China. A total of 880 nasal swabs were randomly collected from swine farms in these regions. The samples were obtained from 5 to 9-month-old fattening pigs, sows, weaning pigs, nursery pigs, and boars exhibiting suspicious clinical symptoms. To identify positive samples, the following procedure was followed: the samples were inoculated into Madin-Darby Canine Kidney (MDCK) cells and cultured at 37 °C for 72 h. The resulting supernatants were collected and subjected to hemagglutination tests and hemagglutination inhibition (HI) tests for subtype determination. The positive samples were then stored at −80 °C for further analysis.
2.2. Cells, mice and pigs
The laboratory stored MDCK cells, porcine alveolar macrophage (3D4/21) cells, and human lung adenocarcinoma epithelial (A549) cells. MDCK cells and A549 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Grand Island, NY, USA), supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 1% penicillin–streptomycin (PS) (BI, Kibbutz Beit Haemek, Israel). 3D4-21 cells were cultured in RPMI-1640 medium containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 1% penicillin–streptomycin (PS), 0.1 mmol/L NEAA, 10 mmol/L Hepes, 2 mmol/L L-GLU, and 2.2 g/L glucose. All cells were maintained at 37 °C with 5% CO2.
We purchased 6-week-old female specific pathogen-free (SPF) BALB/C mice from Liaoning Changsheng Biotechnology Co. Ltd. Additionally, 4-week-old pigs, confirmed serologically negative for influenza, brucellosis, and pseudorabies through hemagglutinin inhibition (HI) and enzyme-linked immunosorbent (ELISA) assays, were acquired from a farm in Zhaoqing City, Guangdong Province.
2.3. Genomic sequencing and phylogenetic analysis
Total RNA was extracted, followed by RT-PCR and PCR using Uni12 and universal primers as described by Hoffmann et al. (2001). The PCR products were subjected to 1% agarose gel electrophoresis, purified, and recovered. Subsequently, the products were cloned into the pMD-19T vector (Takara Bio Inc., Beijing, China) and transformed into DH5α competent cells. The positive recombinant plasmids were screened and sent to Tianyi Huiyuan Gene Technology Co. in Guangdong Province, China, for sequencing.
All reference sequences, including those of H1N1 and H1N2 swine influenza viruses in China, were obtained from the Global Initiative of Sharing All Influenza Data (GISAID). The whole gene sequences of swine influenza viruses were subjected to online BLSAT analysis. Seqman (Version 7.1) and Editseq (DNA Star Lasergene) were utilized for the analysis of sequencing data. Sequence format conversion and amino acid site analysis were performed using MEGA (7.0) software.
For sequence alignment of the eight genes, MAFFT (Version 7.149) was employed, and Model Finder was used to determine the best-fitting model. Based on the AIC criterion, the GTR + F + G4 model (Letunic and Bork, 2021) was selected as the most suitable. The maximum likelihood method available in IQ-TREE (Version 1.68) (Nguyen et al., 2015) was utilized to construct the phylogenetic tree. PhyloSuite Software (Version 1.2.2) was employed for drawing the maximum likelihood (ML) tree using the Bootstrap Ultrafast model with a Bootstrap value of 10,000. Each gene was assigned to an evolutionary lineage (Rambaut et al., 2018). Finally, the evolutionary tree was annotated and enhanced using ITOL (Version 5) (Letunic and Bork, 2021).
2.4. Estimating substitution rates
To assess the genetic distance of the root tips in the ML tree, we employed TempEst, a tool known for its robust temporal signal (Rambaut et al., 2016). For estimating the nucleotide substitution rates of all eight segments, we utilized the Bayesian Markov Chain Monte Carlo (MCMC) method implemented in the BEAST package (version 1.8.2). A coalescent Bayesian Skyline model and a relaxed molecular clock model with uncorrelated log-normally distributed rates were applied. Each set underwent 200 million total steps, with sampling every 1000 steps (Supplementary Table S1). Parameters were evaluated using Tracer (Version 1.6). The posterior distribution of trees obtained from the BEAST analysis was utilized to generate the Bayesian maximum-clade-credibility (MCC) tree for the HA genes (Suchard et al., 2018).
2.5. Virtual replication in vitro
To assess the in vitro growth characteristics of the virus, MDCK cells were employed for influenza virus isolation, while 3D4/21 and A549 cells were also included in the study. When the cell confluence in 12-well plates reached approximately 90%, MDCK cells were infected with the corresponding virus at a multiplicity of infection (MOI) of 0.001. Similarly, 3D4/21 and A549 cells were inoculated with the respective virus at an MOI of 0.01. After a 2-h post-infection (hpi) period, the inoculants were discarded, and the cells were washed twice with PBS. Subsequently, the cells were cultured in Opti-MEM I Reduced Serum Medium (Sigma-Aldrich, St. Louis, MO) supplemented with 1.0 μg/mL TPCK-treated trypsin and 1% penicillin and streptomycin. The supernatants of the cells were collected at 12, 24, 36, 48, 60, and 72 hpi, and titration was performed in MDCK cells. Viral titers were calculated using the Reed-Muench method. The cell growth curve was plotted using GraphPad Prism 7.0 software.
2.6. Receptor-binding assay
The receptor binding characteristics of five H1N2 swIAVs were determined. About 100 μL of streptavidin (PueiMag Biotech, Xiamen, China) with a concentration of 10 μg/mL was added into the 96-well plates and placed at 37 °C for 24 h until dry. The plates were washed three times with phosphate buffered saline containing 0.05 % Tween-20 (PBST), and 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50, 100 ng/100 μL of α-2,3-sialoglycopolymer or α-2,6-sialoglycopolymer (GlycoTech, Gaithersburg, Maryland, USA) were added, respectively, and incubated at 4 °C for 24 h. Then, the plates were washed three times with PBS and incubated with 64 HA units/100 μL H1N2 swIAVs at 4 °C for 12 h. The plates were washed three times with PBST, and then incubated with 0.4 nmol/mL anti-H1 subtype swIAVs monoclonal antibody at 4 °C for 5 h. The plates were washed three times with PBST and incubated with 100 ng/mL horseradish peroxidase (HRP)-labeled goat anti-mouse antibody (Bioworld Technology, Nanjing, China) at 4 °C for 2 h. The plates were washed three times with PBST, then were added 100 μL of 3,3′, 5,5′-tetramethylbenzidine (TMB, Solarbio, Beijing, China), and incubated at room temperature for 5 min. Then, 50 μL of H2SO4 was added to the plates, and the OD450 values were detected using a microplate reader.
2.7. Pathogenicity of H1N2 SIVs to mice
To assess viral replication in mice, a total of 30 specific pathogen free (SPF) BALB/c mice were divided equally into six groups. Five of these groups were intranasally inoculated with the corresponding viruses at a dose of 106 TCID50 in 50 μL under anesthesia. The remaining group received an intranasal inoculation of 50 μL of PBS as a negative control. At 3 days post-infection (dpi), three mice from each infected group were necropsied, and samples including turbinate, trachea, spleen, kidney, brain, and lung were collected. The viral titers in the tissues were determined by titration in MDCK cells. The mice were sacrificed at 14 dpi, and their sera were collected for cross-reactive HI.
2.8. Viral replication and transmission among pigs
To investigate the pathogenicity of the H1N2 subtype swine influenza virus circulating in China, a total of thirteen 4-week-old pigs, confirmed to be negative for influenza virus, were randomly assigned to three groups. Five pigs were intranasally inoculated with SW/198/20 at a dose of 106 TCID50 in 1 mL, constituting the treatment group (Pig 1, Pig 2, Pig 3, Pig 4, and Pig 5). Three pigs received intranasal inoculation with 1 mL of PBS as the control group (Pig 11, Pig 12, and Pig 13). At 1 day post-inoculation (dpi), an additional five pigs were housed together with the treatment group as the physical contact group (Pig 6, Pig 7, Pig 8, Pig 9, and Pig 10). The pigs were observed for symptoms up to 21 dpi. Nasal washes were collected at 1, 3, 5, 7, and 9 dpi, and the viral titers in the washes were determined by titration in MDCK cells. Sera were collected at 7, 14, and 21 dpi, and HI titers were measured using the HI assay with 0.5% turkey erythrocytes. At 3 dpi, two infected pigs, one physical contact pig, and one control pig were euthanized. At 5 dpi, one infected pig, two contact pigs, and one control pig were euthanized. Turbinate, trachea, and lung tissues were collected, and viral titers were determined by titration in MDCK cells. The remaining pigs were euthanized at 21 dpi, and sera were collected.
2.9. Statistic analysis
Statistical analyses were performed using multiple t-test. The “∗” represents significance of difference, data presented as ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
3. Results
3.1. Virus isolation and identification
Viruses were isolated from nasal swab samples from Guangdong, Henan, and Shandong provinces. Isolations were analyzed by using the reverse-transcription polymerase chain reaction (RT-PCR), hemagglutination test, and HI test and confirmed by genomic sequencing and nucleotide BLAST analysis. Based on the results, those isolates were identified as swine influenza A (H1N2) and named A/swine/Shandong/188/2020 (SW/188/20), A/swine/Shandong/198/2020 (SW/198/20), A/swine/Shandong/209/2020 (SW/209/20), A/swine/Maoming/510/2021 (SW/510/21), and A/swine/Henan/700/2021 (SW/700/21). All the whole genomes obtained in this study were deposited to the GISAID's EpiFlu Database under accession numbers: EPI18366359–EPI18366360, EPI18366362–EPI18366364.
3.2. Homology analysis of HA and NA genes
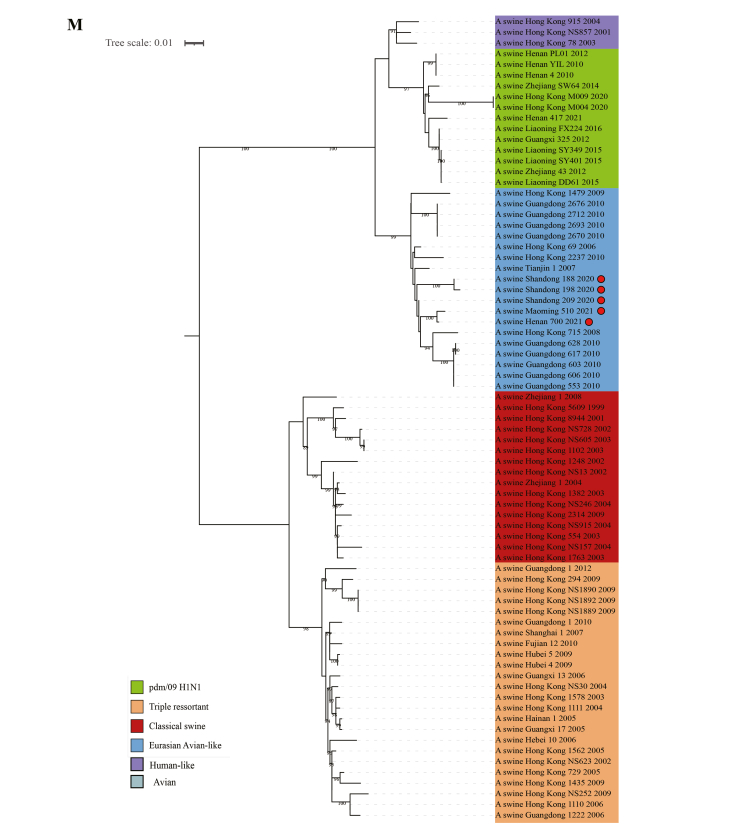
To understand whether the swine H1N2 isolates from this study are related to the previous H1N2 viruses, the homology was determined by comparison with the sequences available in GenBank. The results revealed that the HA genes of the five swH1N2 isolates shared a high degree of similarity (92.48%–99.82%) at the nucleotide level and belonged to the Eurasian avian-like lineage (Supplementary Table S2 and Supplementary Fig. S1).
Among the five viruses, SW/188/20, SW/209/20, SW/510/21, and SW/700/21 showed close similarity to swH1N2 viruses isolated in Liaoning Province in 2014, as indicated by their HA genes. On the other hand, the HA gene of SW/198/20 formed a separate branch and exhibited a close relationship to swH1N2 viruses isolated in Guangdong in 2011 (Fig. 1 and Supplementary Table S2).
Fig. 1.
Phylogenetic relationship of HA gene of swine H1 subtype influenza viruses in China. Different clades are denoted by different markers. All branch lengths are scaled according to the numbers of substitutions per site (subs/site). Phylogenetic tree was estimated using genetic distances calculated by maximum likelihood under the GTR + F + G4 model. SwH1N2 viruses isolated in this study are marked in red.
Regarding the NA genes, the five swH1N2 viruses exhibited nucleotide similarities ranging from 96.88% to 99.79%, indicating their classification within the recent human H3N2 lineage. SW/188/20 and SW/198/20 were found to have high homology with A/Guangdong/Yuefang277/2017/H3N2 (Yuefang277/2017), which is known to cause human infection. SW/209/20 and SW/700/21 showed a close relationship to swH3N2 viruses isolated in Hong Kong in 2014, while SW/510/21 was closely related to swH3N2 viruses isolated in Guangdong Province in 2014 (Fig. 2 and Supplementary Table S2).
Fig. 2.
Phylogenetic trees of the NA gene of influenza A viruses. The viruses marked with red color are the swine H1N2 viruses isolated and sequenced in this study. The most closely related human virus is marked with black circle.
3.3. Homology analysis of internal genes
The homology analysis of the internal genes of the five isolates yielded the following results: PB2 (97.41%–99.60%); PB1 (97.19%–99.96%); NP (97.26%–99.93%); PA (96.79%–99.91%); NS (97.49%–100%); M (96.44%–99.70%) (Supplementary Table S2). Among these, the PB2, NP, and NS genes of SW/188/20, as well as the PB2, PB1, and NP genes of SW/198/20, exhibited the highest similarity, exceeding 99%, to Yuefang277/2017, which was isolated from humans. Furthermore, the M gene of SW/510/21 showed the highest sequence similarity of 99.81% with another virus isolated from humans, namely A/Jiangsu/ALS1/2011/H1N1. The M gene of SW/188/20 displayed a similarity of 98.47% with the corresponding gene of a virus from Hong Kong, while the M gene of SW/198/20 showed a similarity of 97.86% (Supplementary Table S3). Regarding the PB1 and PA genes, SW/188/20, SW/209/20, SW/510/21, and SW/700/21 exhibited the highest homology with two swH1N1 viruses isolated in Liaoning Province in 2014, respectively. As for the PB2 and NS genes, SW/209/20, SW/510/21, and SW/700/21 displayed the highest homology with A/swine/Guangxi/NNXD2023/2013/H1N1 (Supplementary Table S3 and Supplementary Fig. S1).
3.4. Phylogenetic analysis of the viruses
To understand the genetic origin of those viruses' gene segments more precisely, eight phylogenetic trees were constructed using the nucleotide sequences of the five swine H1N2 viruses and reference strains from the GenBank. The phylogenetic analysis results showed that the PB2, PB1, NP, and NS genes of those H1N2 viruses fell into the pdm/09 H1N1 lineage. The HA and M genes of these viruses belonged to the Eurasian Avian-like lineage. The NS genes fell into the triple reassortant lineage (Fig. 3D and Supplementary Fig. S1). The NA gene of the five viruses were special and segregated into current human H3N2 lineage, and they have the closest relationship with human influenza virus Yuefang277/2017 (Fig. 2).
Fig. 3.
Time-scaled evolution of HA gene of swH1N2 viruses. A Analysis of root-to-tip divergence against sampling date for HA gene segment. B A Bayesian Skyride analysis of HA gene of swH1N2 viruses to display changes in the effective population size over time. The solid blue line indicates the median value, and the shaded blue area represents the 95% highest posterior density of genetic diversity estimates. C An maximum-clade-credibility (MCC) tree of the HA sequence of swH1N2 viruses sampled in China is shown, the red marker is swH1N2 virus isolated in this study. D Putative genomic compositions of the swine influenza (H1N2) viruses in this study. The eight genes of influenza virus were represented by horizontal strips, from top to bottom, PB2, PB1, PA, HA, NP, NA, M, NS. Each different color represents a different origin.
3.5. HA gene exhibits the fastest evolutionary rate
To elucidate the evolution of the H1N2 swine influenza virus, we conducted a root-to-tip regression analysis of the time structure for the virus, which exhibited a strong correlation coefficient of 0.89 and an R2 value of 0.95, indicating robust temporal signals suitable for demographic history analysis (Fig. 3A). Notably, there was a significant population expansion of the SWH1N2 virus between 2002 and 2010, as depicted in Fig. 3B.
Furthermore, Bayesian MCC trees were generated based on eight genes, and the evolutionary rates for each gene were estimated (Fig. 3C). The calculated evolutionary rate for the HA gene was 3.1493 × 10−3 substitutions/site/year (95% credibility interval: 2.2934 × 10−3 to 4.0633 × 10−3), while for the NA gene, it was 2.8273 × 10−3 substitutions/site/year (95% credibility interval: 2.1926 × 10−3 to 3.4588 × 10−3). Among the internal genes, the PB2 gene exhibited the fastest evolutionary rate, estimated at 2.6556 × 10−3 substitutions/site/year (95% credibility interval: 1.8416 × 10−3 to 3.3501 × 10−3). On the other hand, the M gene displayed the slowest evolutionary rate, with an estimated rate of 1.715 × 10−3 substitutions/site/year, ranging from 1.1439 × 10−3 to 2.2685 × 10−3 (Fig. 4).
Fig. 4.
Nucleotide substitution rates for each of the eight segments. That shown as mean substitution rate for each gene, with the 95% lower and upper HPD values presented as error bars.
3.6. The five swH1N2 viruses obtained adaptation mutations
To gain deeper insights into the molecular characteristics of those five strains, their whole genomes were analyzed. In the HA genes, the alkaline cleavage site of the H1N2 swine influenza viruses consisted of the sequence REQAG. Furthermore, we detected the presence of 190D and 225E mutations, indicating a strong adaptive ability to mammalian hosts. In the NA genes, we observed the N369K mutation, associated with adaptive changes in human influenza viruses.
The A271S mutation was identified in PB2 genes, which is known to impact the polymerase activity of influenza viruses. Additionally, we found the A676T, L89V, I495V, and T271A mutations (Yuan et al., 2018; Younis et al., 2018; Deleu et al., 2018). In the PA genes, the P224S mutation, associated with influenza virus pathogenicity, was confirmed in all five strains. In the NP gene, we observed a Q357K mutation that potentially enhances the adaptability of swine influenza viruses to the host population, as well as a D53E mutation related to host adaptability (Zhu et al., 2019).
Examining the M gene, we discovered the T215A and S31N mutations, which have implications for pathogenicity and resistance to adamantane, respectively (Mor et al., 2016). In the NS gene, we confirmed the presence of the replication-associated P42S mutation and the K186E mutation in all five strains, which are known to inhibit interferon production (Table 1).
Table 1.
Mutations of swH1N2 viruses isolated in this study.
| Segment | Position | SW/188/20 | SW/198/20 | SW/209/20 | SW/700/21 | SW/510/21 | Mutation |
|---|---|---|---|---|---|---|---|
| PB2 | 89 | V | V | V | V | V | L—V |
| 271 | A | A | A | A | A | T—A | |
| 333 | T | T | T | T | T | T—I | |
| 495 | V | V | V | V | V | I—V | |
| 627 | E | E | E | E | E | E—K | |
| 676 | T | T | T | T | T | A—T | |
| PA | 70 | A | A | A | A | A | A—V |
| 103 | P | P | P | P | P | P—H | |
| 224 | S | S | S | S | S | P—S | |
| 343 | A | A | A | A | A | A—T | |
| 573 | I | I | I | I | I | I—V | |
| 659 | S | S | S | S | S | S—L | |
| HA | 145 | N | N | N | N | N | N—K |
| 158 | G | G | G | G | G | G—N | |
| 190 | D | D | D | D | D | D—V | |
| 225 | E | E | E | D | D | N/G—D/E | |
| 226 | I | I | I | I | I | Q—I | |
| 228 | S | S | S | S | S | Q—S | |
| NP | 34 | G | G | G | G | G | G—A |
| 53 | E | E | E | E | E | D—E | |
| 313 | V | V | V | V | V | V—I | |
| 319 | N | N | N | N | N | N—K | |
| 357 | K | K | K | K | K | Q—K | |
| 479 | L | L | L | L | L | L—F | |
| NA | 119 | E | E | E | E | E | E—V |
| 151 | D | D | D | D | D | D—N | |
| 152 | R | R | R | R | R | R—K | |
| 293 | R | R | R | R | R | R—K | |
| 295 | N | N | N | N | N | N—S | |
| 369 | K | K | K | K | K | N—K | |
| M1 | 26 | L | L | L | L | L | L—F |
| 30 | S | S | S | S | S | S—D | |
| 215 | A | A | A | A | A | T—A | |
| M2 | 30 | A | A | A | A | A | A—P |
| 31 | N | N | N | N | N | S—N | |
| 34 | G | G | G | G | G | G—E | |
| NS | 38 | R | R | R | R | R | R—A |
| 41 | K | K | K | K | K | K—E | |
| 42 | S | S | S | S | S | P—S | |
| 103 | F | F | F | F | F | F—L | |
| 106 | M | M | M | M | M | M—I | |
| 186 | E | E | E | E | E | K—E |
Note: Mutations marked in bold indicate the mutations in strains isolated in this study.
3.7. All the viruses effectively replicated in mammalian cells
To evaluate the replication capability of SW/700/21, SW/188/20, SW/198/20, SW/209/20, and SW/510/21 in vitro, the growth kinetics of all isolated and purified strains were determined in three cell types. Efficient replication was observed for all viruses in MDCK, 3D4/21, and A549 cells. Specifically, the viruses demonstrated efficient replication in 3D4/21 cells, with viral titers ranging from 0 to 9.5 lgTCID50/mL. The maximum titer of 7.53 lgTCID50/mL was reached at 36 hpi, while at 36, 48, and 60 hpi, the titer reached 7.6 lgTCID50/mL (Fig. 5A). Notably, the peak titer of SW/209/20 in 3D4/21 cells surpassed that of SW/700/21, SW/510/21, SW/188/20, and SW/198/20 (P < 0.01).
Fig. 5.
Growth kinetics of five swH1N2 viruses in mammalian cells. 3D4/21 (A) and A549 (B) cells were inoculated with viruses at an MOI of 0.01, and MDCK (C) cells were inoculated with viruses at an MOI of 0.001. The supernatants of cells were collected at 12, 24, 36, 48, 60 and 72 h post-infection and titrated in MDCK cells. Data presented as mean ± S.D. Statistical analyses were performed using multiple t-test. ∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05.
In A549 cells, the virus titers for SW/510/21, SW/209/20, SW/198/20, SW/188/20, and SW/700/21 ranged from 0 to 4.5 lgTCID50/mL. The maximum titer of 4.23 lgTCID50/mL was achieved at 36, 60, and 72 hpi, respectively. Notably, the peak titer of SW/510/21 in A549 cells surpassed that of SW/700/21, SW/188/20, SW/198/20, and SW/209/20 (P < 0.01) (Fig. 5B).
In MDCK cells, the viral titers for SW/700/21, SW/188/20, SW/510/21, SW/209/20, and SW/198/20 ranged from 0 to 8.0 lgTCID50/mL. The maximum titers were achieved at 24, 60, and 72 hpi, respectively. Importantly, the peak titer of SW/700/21 in MDCK cells was higher than that of SW/510/21, SW/188/20, SW/198/20, and SW/209/20 (P < 0.01) (Fig. 5C).
In summary, the replication capabilities of SW/700/21, SW/188/20, SW/198/20, SW/209/20, and SW/510/21 were assessed in vitro using different cell lines. The results indicate variations in peak titers among the strains, with SW/209/20 displaying higher titers in 3D4/21 cells, while SW/510/21 exhibited higher titers in A549 cells. Additionally, SW/700/21 demonstrated a higher peak titer in MDCK cells compared to the other strains (P < 0.01) (Fig. 5).
3.8. Five swH1N2 viruses preferentially bound to the α-2,6 SA-linked receptor
To evaluate the receptor-binding preferences of H1N2 swIAVs, a solid-phase binding assay was performed with two different glycopolymers, α-2,6-siaylglycopolymer (human-like receptor) and α-2,3-siaylglycopolymer (avian-like receptor). The results showed the five SIVs bound SA α 2,6 Gal receptors with high affinity but bound poorly to SAα2,3Gal receptors (Fig. 6).
Fig. 6.
Receptor binding features of five swH1N2 viruses. The binding ability of five swH1N2 viruses to two different biotinylated glycans (30-sialyl-N-acetyllactosamine, 30-SLN, shown in blue; 60-sialyl-N-acetyllactosamine, 60-SLN, shown in red) were detected, and viruses were analyzed at the concentration of 0.78, 1.56, 3.125, 6.25, 12.5, 25, 50 and 100 ng/100 μL. The absorbance value was read at 450 nm. Data presented as mean ± S.D.
3.9. Replication and pathogenicity of the viruses in mice
To investigate the pathogenicity of the H1N2 virus in mice, all five viruses were chosen for an infection experiment. From the second day, mice in the SW/209/20-infected group displayed clinical symptoms, including lethargy, untidy coats, huddling, reluctance to move, and trembling. By the fourth day, there was an improvement in the symptoms.
On the third day post-infection, three mice were randomly euthanized in each group, and their tissues were collected for virus titration. All viruses exhibited efficient replication in the nasal turbinate, trachea, and lung tissues of the inoculated mice. The viral titers in the lung tissues were higher than other tissues, ranging from 0.33 lgEID50/g/mL to 7.5 lgEIID50/g/mL. Viral shedding titers in the trachea tissues ranged from 2.2 lgEID50/g/mL to 3.37 lgEID50/g/mL, while in the turbinate tissues, the range was from 3.5 lgEID50/g/mL to 4.83 lgEID50/g/mL.
Except for SW/188/20, the remaining viruses were notably detected in the kidneys, with titers ranging from 1.5 lgEID50/g/mL to 2.43 lgEID50/g/mL. Additionally, SW/198/20 and SW/209/20 were detected in the spleen, with viral titers of 1.1 lgEID50/g/mL and 1.0 lgEID50/g/mL, respectively. No virus was detected in the brain. All negative controls tested negative for virus shedding in the lung, nasal turbinate, and other tissues (Fig. 7).
Fig. 7.
Pathogenicity of the swH1N2 viruses in mice. Thirty SPF BALB/c mice were equally divided into 6 groups, and 5 groups were intranasally inoculated with corresponding viruses at a dose of 106 TCID50 in 50 μL under anesthesia. The other group was intranasally inoculated with PBS in a volume of 50 μL as a negative control. Three mice in each group were euthanized at day 3 post-infection. Trachea, turbinate, lung, spleen and kidney were collected and the viral titers were titrated in MDCK cells. Virus titers were expressed as lgTCID50/g/mL.
The mice were euthanized at 14 dpi, and serum samples were collected for further analysis. The antibody titers were determined using the HI and Cross-HI tests. The HI results indicated the presence of antibodies in the serum, with antibody titers ranging from 1:20 to 1:640 (Table 2). The antigenicity of the five viruses exhibited some level of variation (Table 3). Notably, SW/510/21 demonstrated significant antigenic differences compared to SW/700/21, SW/209/20, and SW/198/20. Additionally, SW/700/21 displayed significant antigenic differences when compared to SW/209/20 and SW/198/20.
Table 2.
HI titers of sera collected from mice.
| Virus | Treatment |
||||
|---|---|---|---|---|---|
| M1a | M2 | M3 | M4 | M5 | |
| SW/510/21 | 320 | 160 | 320 | 160 | 320 |
| SW/700/21 | 160 | 640 | 160 | 640 | 320 |
| SW/209/20 | 20 | 160 | 50 | 80 | 40 |
| SW/188/20 | 20 | 50 | 100 | 25 | 50 |
| SW/198/20 | 50 | 100 | 25 | 100 | 100 |
| PBS | <10 | <10 | <10 | <10 | <10 |
Means one of the mouse in the each group. HI titers were expressed at geometric mean titer.
Table 3.
Antigenic relatedness (R-value) among the five viruses as determined by the degree of cross-HI activity.
|
R-value |
|||||
|---|---|---|---|---|---|
| SW/510/21 | SW/700/21 | SW/209/20 | SW/188/20 | SW/198/20 | |
| SW/510/21 | 1.000 | 0.500 | 0.559 | 0.707 | 0.500 |
| SW/700/21 | 0.500 | 1.000 | 0.500 | 1.000 | 0.632 |
| SW/209/20 | 0.559 | 0.500 | 1.000 | 0.884 | 0.707 |
| SW/188/20 | 0.707 | 1.000 | 0.884 | 1.000 | 1.000 |
| SW/198/20 | 0.500 | 0.632 | 0.707 | 1.000 | 1.000 |
Notes: R-values shown in bold indicate significant differences. R = 1 indicates the two viruses pose the same antigenicity, 0.67 ≤ R < 1 indicates no significant antigenic difference between the two viruses, and R < 0.5 indicates a significant antigenic difference between the viruses.
3.10. H1N2 SIV effectively replicates and transmits among pigs
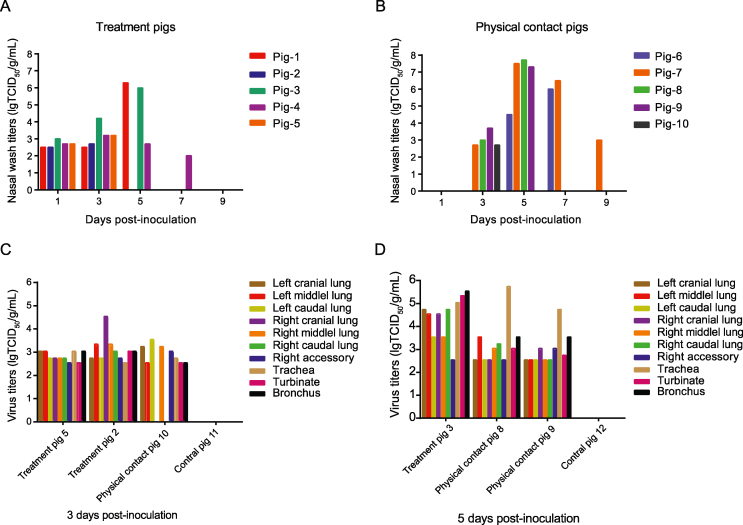
To investigate the infection characteristics of the H1N2 subtype swine influenza virus in pigs, the SW/198/20 strain was selected for the experiment. Five pigs were intranasally inoculated with SW/198 at a dose of 106 TCID50 in 1 mL after being anesthetized. Clinically, the virus-inoculated pigs exhibited cough symptoms. The infected group reached its highest body temperature of 41.8 °C at 3 dpi. Nasal wash samples were collected at 1, 3, 5, 7, and 9 dpi, and the shedding viral titers were titrated in MDCK cells. Virus shedding was detected in both the inoculated and contact group pigs, with peak shedding occurring at 5 dpi with titers of 6.3 lgTCID50/mL and 7.7 lgTCID50/mL, respectively. From day 1 to 7 post-infection, treatment pigs exhibited shedding titers ranging from 2.0 to 6.3 lgTCID50/mL, while contact group pigs showed titers ranging from 2.7 to 7.7 lgTCID50/mL from 3 to 9 dpi. Virus shedding was detected in all pigs from both the treatment and contact groups (Fig. 8A and B).
Fig. 8.
Viral shedding and replication of swH1N2 virus in pigs. Thirteen healthy, 4-week-old pigs with negative sera against influenza viruses were randomly divided into three groups. Of which, five pigs were intranasally inoculated with SW/198 at a dose of 106 TCID50 in 1 mL as the treatment group (A) and were then co-housed with five naïve pigs at day 1 post-infection (dpi) as contact group (B). Three pigs were intranasally inoculated with 1 mL PBS as the control group. Nasal wash was collected from all pigs from 1 to 9 dpi and titrated in MDCK cells. Two treatment pigs, one contact pig and one control pig were euthanized at 3 dpi (C). One treatment pig, two contact pigs and one control pig were euthanized at 5 dpi (D). Lung, trachea and turbinate were collected and the viral titers were titrated in MDCK cells. Virus titers were expressed as lgTCID50/g/mL.
At 3 dpi, two pigs in the treatment group (pigs 2 and 5), one pig in the contact group (pig 10), and one control pig (pig 11) were euthanized to detect viral titers in the respiratory tracts. Virus shedding was observed in respiratory system tissues, including the trachea, turbinate, bronchus, and lung tissues, with titers ranging from 2.5 to 4.5 lgTCID50/g/mL in the treatment group. The highest viral titer of 4.5 lgTCID50/g/mL was detected in the right cranial lung of pig 2. In the cohabitation group, viral titers ranged from 0 to 3.5 lgTCID50/g/mL, with the peak titer of 3.5 lgTCID50/g/mL observed in the left caudal lung. No virus shedding was detected in the right cranial lung, right caudal lung, or any tissues in the contact group or control group (Fig. 8C).
At 5 dpi, one pig from the treatment group (pig 3) and two pigs from the contact group (pig 8, 9) were euthanized, and virus shedding was detected. In the treatment group, viral titers ranged from 2.5 to 5.3 lgTCID50/g/mL, with the turbinate of pig 3 exhibiting the highest titer of 5.3 lgTCID50/g/mL. In the contact group, viral titers ranged from 2.5 to 5.7 lgTCID50/g/mL, with the trachea titer of pig 8 being the highest at 5.7 lgTCID50/g/mL (Fig. 8D).
To determine the seroconversion of swine serum antibodies, swine serum samples were collected at 14 dpi and subjected to HI testing. A HI titer of ≥10 was considered a positive result. All pigs in the infection group and physical contact group showed positive swine serum antibodies, while no seroconversion was detected in the negative control group (Table 4).
Table 4.
HI titers of sera collected from pigs.
| DPI | Treatment |
Contact |
Control |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pig1 | Pig2 | Pig3 | Pig4 | Pig5 | Pig6 | Pig7 | Pig8 | Pig9 | Pig10 | Pig11 | Pig12 | Pig13 | |
| 7 14 |
1280 1280 |
– – |
– – |
160 160 |
– – |
20 20 |
20 20 |
– – |
– – |
– – |
– – |
– – |
<10 <10 |
Note: “-” Indicates that the pig was euthanized at 3 dpi or 5 dpi.
4. Discussion
The first H1N1 subtype avian influenza virus was isolated from a swine influenza pandemic in Belgium in 1979. It is called the Eurasian avian H1N1 swine influenza virus (EAH1N1 SIVs) (Pensaert et al., 1981). Soon after its discovery, EAH1N1 SIVs were detected in pigs in many countries and caused several human infections in Europe and China (de Jong et al., 1988; Myers et al., 2007; Yang et al., 2012; Qi et al., 2013). In China, the EA-like H1N1 SIV was first detected in Hong Kong in 2001. CS, EA, TR, and H3N2 were co-circulated in pig populations from 2002 to 2005, and EA-like H1N1 strains became the dominant lineage in China after 2005 (Tialla et al., 2020; Wang et al., 2022). In this study, the HA and M gene fragments of all isolated strains belonged to the EA branch. Pigs are known as “influenza mixers” where constant recombination of viruses occurs. EA recombinant viruses have been the main EA lineages since their initial detection in Hong Kong in 2007 (Vijaykrishna et al., 2011). Previous studies have shown that EAH1N1 SIVs evolved into five different genotypes through accumulated mutations or the acquisition of different internal gene segments from viruses in other lineages between 2010 and 2013. Researchers have also found that EAH1N1 SIVs formed two distinct antigenic groups, exhibiting markedly different antigenicity from the 2009/H1N1 pandemic virus. Several amino acid changes in PB2 were found to be important for the replication or virulence of influenza viruses in mammals (Gao et al., 2009; Fan et al., 2014; Yamada et al., 2010). It has been confirmed that the PB2 gene of EAH1N1 SIVs has undergone mammalian adaptation mutation (Yang et al., 2016). Since 2009, the pdm/09 H1N1 virus has spread to pig herds worldwide (Weingartl et al., 2010; Pereda et al., 2010). Subsequently, recombination between EA-like H1N1 SIVs and pdm/09 viruses has been found in pigs in China and other countries (Liang et al., 2014; He et al., 2018; Cao et al., 2019; Vijaykrishna et al., 2010).
A large outbreak of a recombinant H1N2 virus occurred in southern Japan during the winter of 1989 and spring of 1990 (Bhatta et al., 2020). Since then, H1N2 has spread widely throughout East Asia. In China, the first report of H1N2 came from Zhejiang Province in 2004 (Pulit-Penaloza et al., 2019). Our study showed a high similarity in the PB2, PB1, NP, NA, and NS genes between strain SW/198/20 and anthropogenic strain Yuefang277/2017. Additionally, there was a high similarity in the PB2, NP, NA, and NS genes between SW/188/20 and Yuefang277/2017. To further analyze the genetic evolution relationship between this individual influenza strain and previous isolated SIVs, we reconstructed the phylogenetic tree using the NA gene. The results showed that the strains from our study appeared after Yuefang277/2017, suggesting that SW/198/20 and SW/188/20 may have acquired some internal genes from Yuefang277/2017. We also found that the PB2, PB1, HA, NA, and NS genes of Yuefang277/2017 had the highest homology with H3N2 swine influenza. This finding provides further evidence that pigs serve as an “influenza virus mixing vessel” and that increased surveillance of swine influenza viruses can help prevent the emergence of human pandemic strains.
It is widely accepted that changes in the position and number of glycosylation sites on the HA protein play a role in the evolution of influenza viruses (Sun et al., 2012). In our study, we observed that among the eight fragments of influenza viruses, the HA gene exhibited the fastest nucleotide substitution rate, while the M gene showed the slowest rate. This finding aligns with the fact that the HA gene is often prone to mutations.
In this study, we isolated and purified five strains of swine influenza viruses, which effectively replicate in MDCK, A549, and 3D4/21 cells. The viral titers of all strains tended to be average on 3D4/21 cells, while the growth of SW/700/21 and SW/510/21 was significantly better than that of other strains in A549 and MDCK cells. Previous studies have indicated that the presence of the G78D mutation in the HA protein contributes to the growth of the H3N2 human seasonal influenza virus in cells (Lee et al., 2018; Anderson et al., 2016). Interestingly, SW/700/21 and SW/510/21 in our study possessed this mutation, and both viruses exhibited a higher replicative ability in MDCK cells than other strains. Further investigation is required to determine if these mutations are indeed responsible for this phenomenon. The PB2 protein is known to play a critical role in determining the virulence and host adaptability of influenza viruses, and we identified the presence of the 590S mutation in the PB2 protein in all of our strains (Li et al., 2005; Manz et al., 2013; Pulit-Penaloza et al., 2019). This change may be associated with the virus's ability to replicate in cells.
The pathogenicity of the viruses in mice was assessed, revealing that the viruses caused varying degrees of weight loss and demonstrated effective replication in the respiratory organs. Additionally, the results of this study indicated that some viruses could replicate effectively in the spleen and kidney. A recent study found that substituting S328L at the hemagglutinin cleavage site of an EA-like H1N2 swine influenza virus reduced weight loss and increased survival in mice (Cai et al., 2021; Jung et al., 2020). In our study, all five SIVs had the S328L mutation at the HA cleavage sites, which may explain why the isolated strains exhibited lower pathogenicity. Our strains demonstrated a 100% replication rate in the lungs, trachea, and turbinate, consistent with previous findings. SW/198/20 and SW/209/20 showed effective replication in the spleen, albeit with lower replication titers. Except for SW/188/20, all strains were detectable in the kidney. Previous studies have demonstrated the effective replication of SIVs in the kidneys. Mammalian adaptive mutations 591Q and 701N in the PB2 protein were found in all five SIV strains in this study, which may be associated with the robust replication of the swine influenza viruses in mice.
Swine H1N2 influenza viruses carrying avian-origin HA and 2009 pandemic PA and NP genes exhibited higher virulence in pigs and mice (Jang et al., 2020). Based on the analysis above, the PB2, PB1, NP, NA, and NS genes of SW/198/20 showed high homology with human influenza strains. Consequently, SW/198/20 was selected to conduct pathogenicity tests in pigs. Following infection with the SW/198/20 strain, pigs exhibited symptoms such as sneezing and a runny nose, with the highest recorded temperature reaching 41.8 °C as measured daily. Virus titers were detected in the nasal wash and organs of both infected and cohabitating pigs, indicating that SW/198/20 replicates effectively and has high transmissibility in pigs, with a transmission rate of 100%. At 14 days post-infection (dpi), both the contact group and the treatment group tested positive in their serum samples. In both the contact and treatment groups, viral shedding peaked at 5 dpi, with variable virus titers observed in different lung regions. These findings contribute to enhancing our understanding of swine influenza viruses.
In this study, we evaluated pathogenicity of viruses mainly according to their replication and transmission ability in pigs. The pathology of tissues will be considered in the future studies. The G228S mutation in the HA protein facilitates efficient replication and transmission of swine influenza viruses in pigs (Ma et al., 2015). Notably, the SW/198/20 virus possesses the 228S mutation in the HA protein and the 53E mutation in the NP gene, which may explain its ease of replication and transmission in pigs. The potential glycosylation sites of the five swine influenza viruses were analyzed. All strains shared the following five glycosylation sites (28NST30, 40NVT42, 291NCT293, 498NGT500, 557NGS559), while SW/198/20 had an additional glycosylation site at 502NYS504 compared to the other strains. The increase in glycosylation sites may be related to the strain's heightened pathogenicity (Bourret, 2018; Mon et al., 2020).
5. Conclusions
In this study, we isolated five strains of quadruple reassortant H1N2 viruses from pig herds. These viruses carry the NA gene from the current human H3N2 lineage, and several genes of these H1N2 swine influenza viruses exhibit high sequence similarity to human influenza virus A/Guangdong/Yue Fang277/2017 (H3N2). Moreover, the HA gene of swH1N2 exhibits a rapid evolutionary rate. The virus replicates effectively in mammalian cells and can efficiently replicate in pigs and mice. Additionally, it readily transmits among pigs, indicating its potential for zoonotic and pandemic spread.
Data availability
All the data generated during the current study are included in the manuscript.
Ethics statement
The laboratory experiments were carried out in compliance with biosafety committee of South China Agriculture University approved protocols. The handling of sampling was performed in accordance with experimental animal administration and ethics committee of South China Agriculture University (SCAUABSL2020-7; 13 March 2021) approved guideline.
Author contributions
Xinxin Cui: investigation, data curation, writing-original draft. Jinhuan Ma: methodology, writing-original draft. Zifeng Pang: Visualization, writing-original draft. Lingzhi Chi: Resources. Cuishan Mai: Software. Hanlin Liu: Methodology. Ming Liao: conceptualization, funding acquisition. Hailiang Sun: conceptualization, writing-review & editing.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank the data submitters from the GISAID and GenBank Flu databases for the influenza A virus sequences. This work was supported by Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science-Distinguished Scholar (R2020PY-JC001).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2024.02.002.
Contributor Information
Ming Liao, Email: mliao@scau.edu.cn.
Hailiang Sun, Email: hsun@scau.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
figs6.
References
- Anderson T.K., Macken C.A., Lewis N.S., Scheuermann R.H., Van Reeth K., Brown I.H., Swenson S.L., Simon G., Saito T., Berhane Y., Ciacci-Zanella J., Pereda A., Davis C.T., Donis R.O., Webby R.J., Vincent A.L. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. mSphere. 2016;1 doi: 10.1128/mSphere.00275-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barman S., Krylov P.S., Fabrizio T.P., Franks J., Turner J.C., Seiler P., Wang D., Rehg J.E., Erickson G.A., Gramer M., Webster R.G., Webby R.J. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J.A., Eckert A.M., Tumpey T.M., Maines T.R. Complexities in ferret influenza virus pathogenesis and transmission models. Microbiol. Mol. Biol. Rev. 2016;80:733–744. doi: 10.1128/MMBR.00022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta T.R., Ryt-Hansen P., Nielsen J.P., Larsen L.E., Larsen I., Chamings A., Goecke N.B., Alexandersen S. Infection dynamics of swine influenza virus in a Danish pig herd reveals recurrent infections with different variants of the H1N2 swine influenza A virus subtype. Viruses-Basel. 2020;12:1013. doi: 10.3390/v12091013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret V. Avian influenza viruses in pigs: an overview. Vet. J. 2018;239:7–14. doi: 10.1016/j.tvjl.2018.07.005. [DOI] [PubMed] [Google Scholar]
- Brown I.H., Harris P.A., Mccauley J.W., Alexander D.J. Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J. Gen. Virol. 1998;79(Pt 12):2947–2955. doi: 10.1099/0022-1317-79-12-2947. [DOI] [PubMed] [Google Scholar]
- Cai M., Zhong R., Qin C., Yu Z., Huang J., Wen X., Ji C., Chen Y., Cai Y., Yi H., Gong L., Zhang G. Ser-Leu substitution at P2 position of the hemagglutinin cleavage site attenuates replication and pathogenicity of Eurasian avian-like H1N2 swine influenza viruses. Vet. Microbiol. 2021;253:108847. doi: 10.1016/j.vetmic.2020.108847. [DOI] [PubMed] [Google Scholar]
- Cao Z., Zeng W., Hao X., Huang J., Cai M., Zhou P., Zhang G. Continuous evolution of influenza A viruses of swine from 2013 to 2015 in Guangdong, China. PLoS One. 2019;14 doi: 10.1371/journal.pone.0217607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chare E.R., Gould E.A., Holmes E.C. Phylogenetic analysis reveals a low rate of homologous recombination in negative-sense RNA viruses. J. Gen. Virol. 2003;84:2691–2703. doi: 10.1099/vir.0.19277-0. [DOI] [PubMed] [Google Scholar]
- De Jong J.C., Paccaud M.F., De Ronde-Verloop F.M., Huffels N.H., Verwei C., Weijers T.F., Bangma P.J., Van Kregten E., Kerckhaert J.A., Wicki F., et al. Isolation of swine-like influenza A(H1N1) viruses from man in Switzerland and The Netherlands. Ann. Inst. Pasteur. Virol. 1988;139:429–437. doi: 10.1016/s0769-2617(88)80078-9. [DOI] [PubMed] [Google Scholar]
- De Jong J.C., Smith D.J., Lapedes A.S., Donatelli I., Campitelli L., Barigazzi G., Van Reeth K., Jones T.C., Rimmelzwaan G.F., Osterhaus A.D., Fouchier R.A. Antigenic and genetic evolution of swine influenza A (H3N2) viruses in Europe. J. Virol. 2007;81:4315–4322. doi: 10.1128/JVI.02458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleu S., Kakuda T.N., Spittaels K., Vercauteren J.J., Hillewaert V., Lwin A., Leopold L., Hoetelmans R.M.W. Single- and multiple-dose pharmacokinetics and safety of pimodivir, a novel, non-nucleoside polymerase basic protein 2 subunit inhibitor of the influenza A virus polymerase complex, and interaction with oseltamivir: a Phase 1 open-label study in healthy volunteers. Br. J. Clin. Pharmacol. 2018;84:2663–2672. doi: 10.1111/bcp.13733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hatta M., Kim J.H., Halfmann P., Imai M., Macken C.A., Le M.Q., Nguyen T., Neumann G., Kawaoka Y. Novel residues in avian influenza virus PB2 protein affect virulence in mammalian hosts. Nat. Commun. 2014;5:5021. doi: 10.1038/ncomms6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Zhang Y., Shinya K., Deng G., Jiang Y., Li Z., Guan Y., Tian G., Li Y., Shi J., Liu L., Zeng X., Bu Z., Xia X., Kawaoka Y., Chen H. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C.Q., Han G.Z., Wang D., Liu W., Li G.R., Liu X.P., Ding N.Z. Homologous recombination evidence in human and swine influenza A viruses. Virology. 2008;380:12–20. doi: 10.1016/j.virol.2008.07.014. [DOI] [PubMed] [Google Scholar]
- He P., Wang G., Mo Y., Yu Q., Xiao X., Yang W., Zhao W., Guo X., Chen Q., He J., Liang M., Zhu J., Ding Y., Wei Z., Ouyang K., Liu F., Jian H., Huang W., Garcia-Sastre A., Chen Y. Novel triple-reassortant influenza viruses in pigs, Guangxi, China. Emerg. Microbes Infect. 2018;7:85. doi: 10.1038/s41426-018-0088-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E., Stech J., Guan Y., Webster R.G., Perez D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- Ito T., Couceiro J.N., Kelm S., Baum L.G., Krauss S., Castrucci M.R., Donatelli I., Kida H., Paulson J.C., Webster R.G., Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y., Seo T., Seo S.H. Higher virulence of swine H1N2 influenza viruses containing avian-origin HA and 2009 pandemic PA and NP in pigs and mice. Arch. Virol. 2020;165:1141–1150. doi: 10.1007/s00705-020-04572-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung O., Radenkovic M., Stojanovic S., Lindner C., Batinic M., Gorke O., Pissarek J., Prohl A., Najman S., Barbeck M. In vitro and in vivo biocompatibility analysis of a new transparent collagen-based wound membrane for tissue regeneration in different clinical indications. In Vivo. 2020;34:2287–2295. doi: 10.21873/invivo.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Ye J., Guan S., Liu J., Pu J. Epidemic status of Swine influenza virus in China. Indian J. Microbiol. 2014;54:3–11. doi: 10.1007/s12088-013-0419-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.M., Huddleston J., Doud M.B., Hooper K.A., Wu N.C., Bedford T., Bloom J.D. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E8276–E8285. doi: 10.1073/pnas.1806133115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Chen H., Jiao P., Deng G., Tian G., Li Y., Hoffmann E., Webster R.G., Matsuoka Y., Yu K. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J. Virol. 2005;79:12058–12064. doi: 10.1128/JVI.79.18.12058-12064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Lam T.T., Fan X., Chen X., Zeng Y., Zhou J., Duan L., Tse M., Chan C.H., Li L., Leung T.Y., Yip C.H., Cheung C.L., Zhou B., Smith D.K., Poon L.L., Peiris M., Guan Y., Zhu H. Expansion of genotypic diversity and establishment of 2009 H1N1 pandemic-origin internal genes in pigs in China. J. Virol. 2014;88:10864–10874. doi: 10.1128/JVI.01327-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Shen H., Liu Q., Bawa B., Qi W., Duff M., Lang Y., Lee J., Yu H., Bai J., Tong G., Hesse R.A., Richt J.A., Ma W. Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J. Virol. 2015;89:2831–2841. doi: 10.1128/JVI.03355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz B., Schwemmle M., Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J. Virol. 2013;87:7200–7209. doi: 10.1128/JVI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mon P.P., Thurain K., Janetanakit T., Nasamran C., Bunpapong N., Aye A.M., San Y.Y., Tun T.N., Amonsin A. Swine influenza viruses and pandemic H1N1-2009 infection in pigs, Myanmar. Transbound. Emerg. Dis. 2020;67:2653–2666. doi: 10.1111/tbed.13616. [DOI] [PubMed] [Google Scholar]
- Mor A., White A., Zhang K., Thompson M., Esparza M., Munoz-Moreno R., Koide K., Lynch K.W., Garcia-Sastre A., Fontoura B.M. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 2016;1:16069. doi: 10.1038/nmicrobiol.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers K.P., Olsen C.W., Gray G.C. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 2007;44:1084–1088. doi: 10.1086/512813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerome K., Yoshioka Y., Sakamoto S., Yasuhara H., Oya A. Characterization of a 1980-swine recombinant influenza virus possessing H1 hemagglutinin and N2 neuraminidase similar to that of the earliest Hong Kong (H3N2) virus. Arch. Virol. 1985;86:197–211. doi: 10.1007/BF01309825. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi A., Nerome K., Kanegae Y., Ishida M., Nerome R., Hayashi K., Hashimoto T., Kaji M., Kaji Y., Inaba Y. Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J. Gen. Virol. 1996;77(Pt 8):1751–1759. doi: 10.1099/0022-1317-77-8-1751. [DOI] [PubMed] [Google Scholar]
- Pensaert M., Ottis K., Vandeputte J., Kaplan M.M., Bachmann P.A. Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull. World Health Organ. 1981;59:75–78. [PMC free article] [PubMed] [Google Scholar]
- Pereda A., Cappuccio J., Quiroga M.A., Baumeister E., Insarralde L., Ibar M., Sanguinetti R., Cannilla M.L., Franzese D., Escobar Cabrera O.E., Craig M.I., Rimondi A., Machuca M., Debenedetti R.T., Zenobi C., Barral L., Balzano R., Capalbo S., Risso A., Perfumo C.J. Pandemic (H1N1) 2009 outbreak on pig farm, Argentina. Emerg. Infect. Dis. 2010;16:304–307. doi: 10.3201/eid1602.091230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza J.A., Belser J.A., Tumpey T.M., Maines T.R. Mammalian pathogenicity and transmissibility of a reassortant Eurasian avian-like A(H1N1v) influenza virus associated with human infection in China (2015) Virology. 2019;537:31–35. doi: 10.1016/j.virol.2019.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Cui L., Jiao Y., Pan Y., Li X., Zu R., Huo X., Wu B., Tang F., Song Y., Zhou M., Wang H., Cardona C.J., Xing Z. Antigenic and genetic characterization of a European avian-like H1N1 swine influenza virus from a boy in China in 2011. Arch. Virol. 2013;158:39–53. doi: 10.1007/s00705-012-1423-7. [DOI] [PubMed] [Google Scholar]
- Rambaut A., Drummond A.J., Xie D., Baele G., Suchard M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Lam T.T., Max Carvalho L., Pybus O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2 doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Li X., Goraya M.U., Wang S., Chen J.L. Evolution of influenza A virus by mutation and re-assortment. Int. J. Mol. Sci. 2017;18:1650. doi: 10.3390/ijms18081650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchard M.A., Lemey P., Baele G., Ayres D.L., Drummond A.J., Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wang Q., Zhao F., Chen W., Li Z. Prediction of biological functions on glycosylation site migrations in human influenza H1N1 viruses. PLoS One. 2012;7 doi: 10.1371/journal.pone.0032119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tialla D., Sausy A., Cisse A., Sagna T., Ilboudo A.K., Ouedraogo G.A., Hubschen J.M., Tarnagda Z., Snoeck C.J. Serological evidence of swine exposure to pandemic H1N1/2009 influenza A virus in Burkina Faso. Vet. Microbiol. 2020;241:108572. doi: 10.1016/j.vetmic.2019.108572. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D., Poon L.L., Zhu H.C., Ma S.K., Li O.T., Cheung C.L., Smith G.J., Peiris J.S., Guan Y. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science. 2010;328:1529. doi: 10.1126/science.1189132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D., Smith G.J., Pybus O.G., Zhu H., Bhatt S., Poon L.L., Riley S., Bahl J., Ma S.K., Cheung C.L., Perera R.A., Chen H., Shortridge K.F., Webby R.J., Webster R.G., Guan Y., Peiris J.S. Long-term evolution and transmission dynamics of swine influenza A virus. Nature. 2011;473:519–522. doi: 10.1038/nature10004. [DOI] [PubMed] [Google Scholar]
- Wang S., Wang M., Yu L., Wang J., Yan J., Rong X., Zhou Y., Shan T., Tong W., Li G., Zheng H., Tong G., Yu H. Genetic characterization and pathogenicity of a reassortant Eurasian avian-like H1N1 swine influenza virus containing an internal gene cassette from 2009 pandemic H1N1 virus. Virol. Sin. 2022;37:627–630. doi: 10.1016/j.virs.2022.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartl H.M., Berhane Y., Hisanaga T., Neufeld J., Kehler H., Emburry-Hyatt C., Hooper-Mcgreevy K., Kasloff S., Dalman B., Bystrom J., Alexandersen S., Li Y., Pasick J. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J. Virol. 2010;84:2245–2256. doi: 10.1128/JVI.02118-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S., Hatta M., Staker B.L., Watanabe S., Imai M., Shinya K., Sakai-Tagawa Y., Ito M., Ozawa M., Watanabe T., Sakabe S., Li C., Kim J.H., Myler P.J., Phan I., Raymond A., Smith E., Stacy R., Nidom C.A., Lank S.M., Wiseman R.W., Bimber B.N., O'connor D.H., Neumann G., Stewart L.J., Kawaoka Y. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Chen Y., Qiao C., He X., Zhou H., Sun Y., Yin H., Meng S., Liu L., Zhang Q., Kong H., Gu C., Li C., Bu Z., Kawaoka Y., Chen H. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:392–397. doi: 10.1073/pnas.1522643113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Qiao C., Tang X., Chen Y., Xin X., Chen H. Human infection from avian-like influenza A (H1N1) viruses in pigs, China. Emerg. Infect. Dis. 2012;18:1144–1146. doi: 10.3201/eid1807.120009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.R., Kuo C.Y., Yu I.L., Kung F.Y., Wu F.T., Lin J.S., Liu M.T. Human infection with a reassortant swine-origin influenza A(H1N2)v virus in Taiwan, 2021. Virol. J. 2022;19:63. doi: 10.1186/s12985-022-01794-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis S., Kamel W., Falkeborn T., Wang H., Yu D., Daniels R., Essand M., Hinkula J., Akusjarvi G., Andersson L. Multiple nuclear-replicating viruses require the stress-induced protein ZC3H11A for efficient growth. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E3808–E3816. doi: 10.1073/pnas.1722333115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., Wen L., Zhou J. Inhibitors of influenza A virus polymerase. ACS Infect. Dis. 2018;4:218–223. doi: 10.1021/acsinfecdis.7b00265. [DOI] [PubMed] [Google Scholar]
- Zhu H., Webby R., Lam T.T., Smith D.K., Peiris J.S., Guan Y. History of Swine influenza viruses in Asia. Curr. Top. Microbiol. Immunol. 2013;370:57–68. doi: 10.1007/82_2011_179. [DOI] [PubMed] [Google Scholar]
- Zhu W., Feng Z., Chen Y., Yang L., Liu J., Li X., Liu S., Zhou L., Wei H., Gao R., Wang D., Shu Y. Mammalian-adaptive mutation NP-Q357K in Eurasian H1N1 Swine Influenza viruses determines the virulence phenotype in mice. Emerg. Microbes Infect. 2019;8:989–999. doi: 10.1080/22221751.2019.1635873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated during the current study are included in the manuscript.