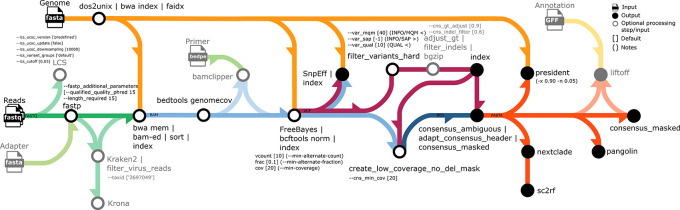
Figure 1. Overview of the CoVpipe2 workflow.
The illustration shows all input (
 ) and output (
) and output (
 ) files as well as optional processing steps and optional input (
) files as well as optional processing steps and optional input (
 ). For each computational step, the used parameters and default values (in brackets [...]) are provided, as well as additional comments ((...)). The arrows connect all steps and are colored to distinguish different data processing steps: green – read (FASTQ) quality control and taxonomy filtering, yellow – reference genome (FASTA) and reference annotation (GFF) for lift-over, blue – mapping files (BAM) and low coverage filter (BED), purple – variant calls (VCF), orange – consensus sequence (FASTA). The icons and diagram components that make up the schematic figure were originally designed by James A. Fellow Yates and
nf-core under a CCO license (public domain).
). For each computational step, the used parameters and default values (in brackets [...]) are provided, as well as additional comments ((...)). The arrows connect all steps and are colored to distinguish different data processing steps: green – read (FASTQ) quality control and taxonomy filtering, yellow – reference genome (FASTA) and reference annotation (GFF) for lift-over, blue – mapping files (BAM) and low coverage filter (BED), purple – variant calls (VCF), orange – consensus sequence (FASTA). The icons and diagram components that make up the schematic figure were originally designed by James A. Fellow Yates and
nf-core under a CCO license (public domain).