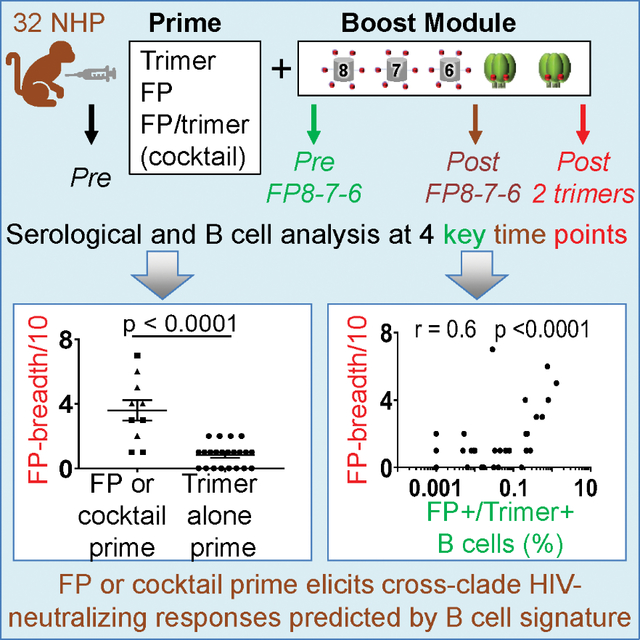
SUMMARY
The HIV fusion peptide (FP) is a promising vaccine target. FP-directed monoclonal antibodies from vaccinated macaques have been identified that neutralize up to ~60% of HIV strains; these vaccinations, however, have involved ~1 year with an extended neutralization-eclipse phase without measurable serum neutralization. Here, in 32 macaques, we test seven vaccination regimens, each comprising multiple immunizations of FP-carrier conjugates and HIV envelope (Env) trimers. Comparisons of vaccine regimens reveal FP-carrier conjugates to imprint cross-clade neutralizing responses and a cocktail of FP conjugate and Env trimer to elicit the earliest broad responses. We identify a signature, appearing as early as week 6 and involving the frequency of B cells recognizing both FP and Env trimer, predictive of vaccine-elicited breadth ~1 year later. Immune monitoring of B cells in response to vaccination can thus enable vaccine insights even in the absence of serum neutralization, here identifying FP imprinting, cocktail approach, and early signature as means to improve FP-directed vaccine responses.
In Brief
Immune monitoring of B cells in response to vaccination can enable early insights, even in the absence of serum neutralization. Cheng et al. observe an early B cell signature in NHPs predictive of the vaccine outcome, with priming of HIV FP imprinting cross-clade neutralizing FP-directed vaccine responses.
Graphical Abstract
INTRODUCTION
The elicitation of protective immune responses against highly diverse viruses remains one of the outstanding challenges of modern vaccinology. Despite decades of research, effective vaccines capable of providing protection against diverse strains of influenza A virus and HIV-1 are still being sought (Erbelding et al., 2018; Kwong and Mascola, 2018). One promising approach is the “antibody-to-vaccine” paradigm in which a broadly neutralizing antibody provides a template to guide the development of immunogens to elicit similar antibodies by vaccination (Burton, 2002; Burton et al., 2004; Crowe, 2016). This approach is attractive because B cell mechanisms of antibody development are well understood and, in principle, can be exploited for optimization.
Recently, for HIV-1, fusion peptide (FP)-directed antibodies (Blattner et al., 2014; Kong et al., 2016; Lee et al., 2016; van Gils et al., 2016; Yuan et al., 2019) have been used as templates for vaccine design, and FP-directed cross-clade neutralizing sera have been elicited in mice, guinea pigs, and non-human primates (NHPs) (Cheng et al., 2019; Kong et al., 2019; Xu et al., 2018). In mice, cross-reactive serum neutralization could be elicited by vaccination in as early as 2–3 months; the most potent titers (Xu et al., 2018) arose after three immunizations with keyhole limpet hemocyanin (KLH)-carrier protein-conjugated peptides corresponding to the N-terminal eight, seven, and six residues of FP, followed by two immunizations with envelope (Env) trimer stabilized in a prefusion-closed state (Kwon et al., 2015; Sanders et al., 2013). In guinea pigs (Cheng et al., 2019; Xu et al., 2018) and NHPs (Kong et al., 2019; Xu et al., 2018), immunization schemes involving this “FP8–7-6-trimer-trimer module” elicited up to 20%–30% neutralization breadth, but development of neutralization was slower, with a “neutralization-eclipse phase” exhibiting no observable serum neutralization against even the most sensitive glycan-deleted viruses for a period lasting upward of 6 months.
In an attempt to understand and improve elicited responses, we vaccinated 32 NHPs with seven vaccine regimens, all of which incorporated FP8–7-6-trimer-trimer as a boosting module. Two of the regimens (in the NHP-1 study) examined the effect of removing glycans neighboring the FP site of vulnerability, similar to glycan-deletion strategies that have been investigated for the CD4-binding site (CD4bs) (Dubrovskaya et al., 2017; Zhou et al., 2017) and the glycan-V3 site (Escolano et al., 2019). Three regimens (in the NHP-2 study) examined the impact of priming with a reagent for dual targeting of FP and CD4bs, and two regimens (in the NHP-3 study) examined the effect of FP priming, either alone or in combination with Env trimer as a priming cocktail. We tracked longitudinally the development of serum neutralization and correlated this with longitudinal characteristics of antigen-specific B cells. Overall, we found effective priming by an FP-coupled carrier to correlate with titer of cross-reactive serum neutralization after completion of the boosting module; by contrast, priming with trimer alone did not induce neutralization breadth. Notably, priming with a cocktail comprising both FP-carrier and Env trimer yielded early cross-reactive responses. Furthermore, we identified a signature related to the frequency of B cells recognizing antigen; this B cell signature could be observed as early as week 6 and correlated with neutralization breadth at the end of the study.
RESULTS
Priming with FP-Proximal Glycan-Deleted Trimers Induces FP-Directed Responses of Low Neutralizing Breadth and Potency (NHP-1 Study)
Glycan shielding of the Env trimer is a key mechanism by which HIV-1 evades the humoral immune system (Lee et al., 2016; Stewart-Jones et al., 2016; Wei et al., 2003). Four potential sites of glycosylation surround FP, at residues N88, N230, N241 and N611, each of which can shield FP from immune recognition (Figure 1A). While BG505 virus lacks a potential site of glycosylation at N241, the transmitted founder virus from the clade C strain of donor CH505 contains all four of the canonical FP-proximal sites of glycosylation (Liao et al., 2013; Saunders et al., 2017). Since the removal of glycans around FP was expected to increase FP-directed immunogenicity, we assessed the impact of priming with two variants of the CH505 Env trimer: (1) “degly3,” with three glycosylation sites at N230, N241, and N611 removed, leaving only N88, which is critical for neutralization by the FP-directed antibody VRC34.01, a vaccine-template antibody from natural infection (Kong et al., 2016); and (2) “degly4,” with all four glycosylation sites neighboring FP removed. These trimers appeared as prefusion-closed Env-trimer structures by electron microscopy (EM) and bound to HIV-neutralizing antibodies similar to wild-type CH505 trimer (Cheng et al., 2019) (Figure 1B).
Figure 1. Immunogen Design and Characterization.
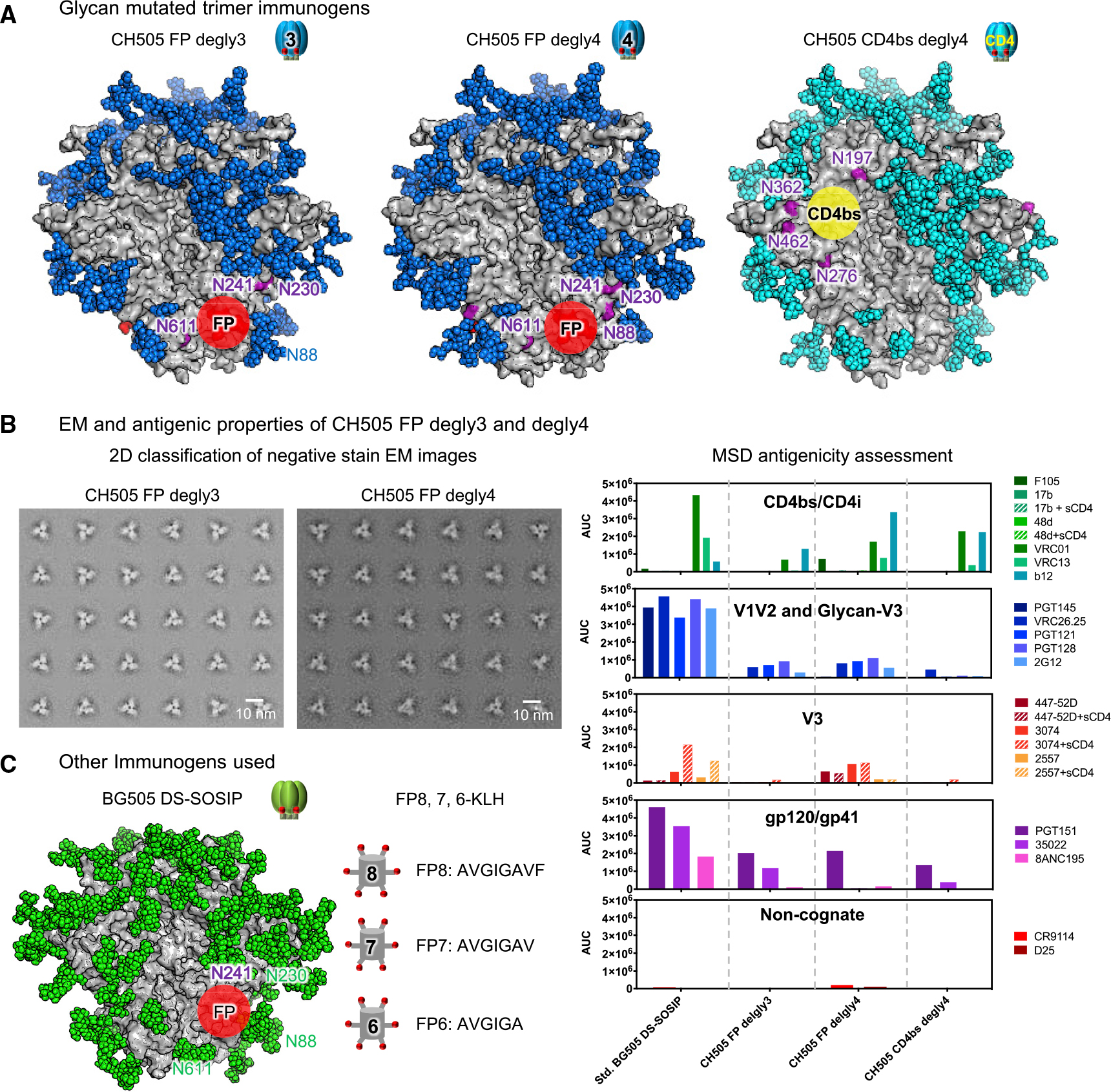
(A) Glycan mutant trimer immunogens. Surface view of the three glycan-deleted CH505 Env-trimer constructs used in this paper. Env protein surface is colored in gray. Deleted N-linked glycans near FP and CD4bs to expose FP or CD4bs are labeled in purple. All other glycans are shown in either blue or cyan to match the immunogen symbols used in other figures.
(B) Physical and antigenic properties of CH505 FP degly3 and degly4.
(C) BG505 Env trimer and FP8–7-6-KLH are shown schematically. Env protein is colored gray, while glycans are colored green.
We immunized two groups of NHPs with either degly3 or degly4 at weeks 0, 4, and 16, followed by a common boosting module (Figure 2A). FP-directed immunoglobulin G (IgG) responses were readily detectable by ELISA after two immunizations (Figure 2B), and degly4 induced significantly higher anti-FP IgG serum responses than degly3 by week 18, prior to the boosting module (“pre FP8–7-6”) (p = 0.0238) (Figure 2C; Table S1), indicating that removal of all glycans around FP enhanced the immunogenicity of the FP site. On the other hand, both groups induced similar responses to wild-type BG505 Env trimer over the entire course of immunization (Figures 2D and 2E; Table S1), suggesting that the higher FP-directed responses induced by degly4 did not translate to higher ELISA responses against wild-type trimers. In terms of neutralization, at week 18 prior to the boosting module (pre FP8–7-6), immune sera failed to neutralize consistently wild-type BG505 or CH505 viruses but could neutralize deglycosylated mutants of BG505 Δ611 (N611Q) and Δ88Δ611 (N88Q+N611Q) (Table S2), which are especially sensitive to neutralization by FP-directed antibodies (Kong et al., 2016; Xu et al., 2018). After the boosting module, titers against Δ611 for both groups of animals increased ~2-fold (Figures 2F and 2G). Overall, at week 66, after two immunizations of BG505 trimer (“post 2 trimers”), all animals generated neutralizing activities against BG505 Δ611 virus; however, only sporadic low-titer neutralizing activity was observed on a 10-wild-type-virus panel (Figure 2G).
Figure 2. Immunizations Using Trimers with Deleted FP-Proximal Glycans Consistently Induce FP-Directed Responses, but with Low Neutralizing Breadth.
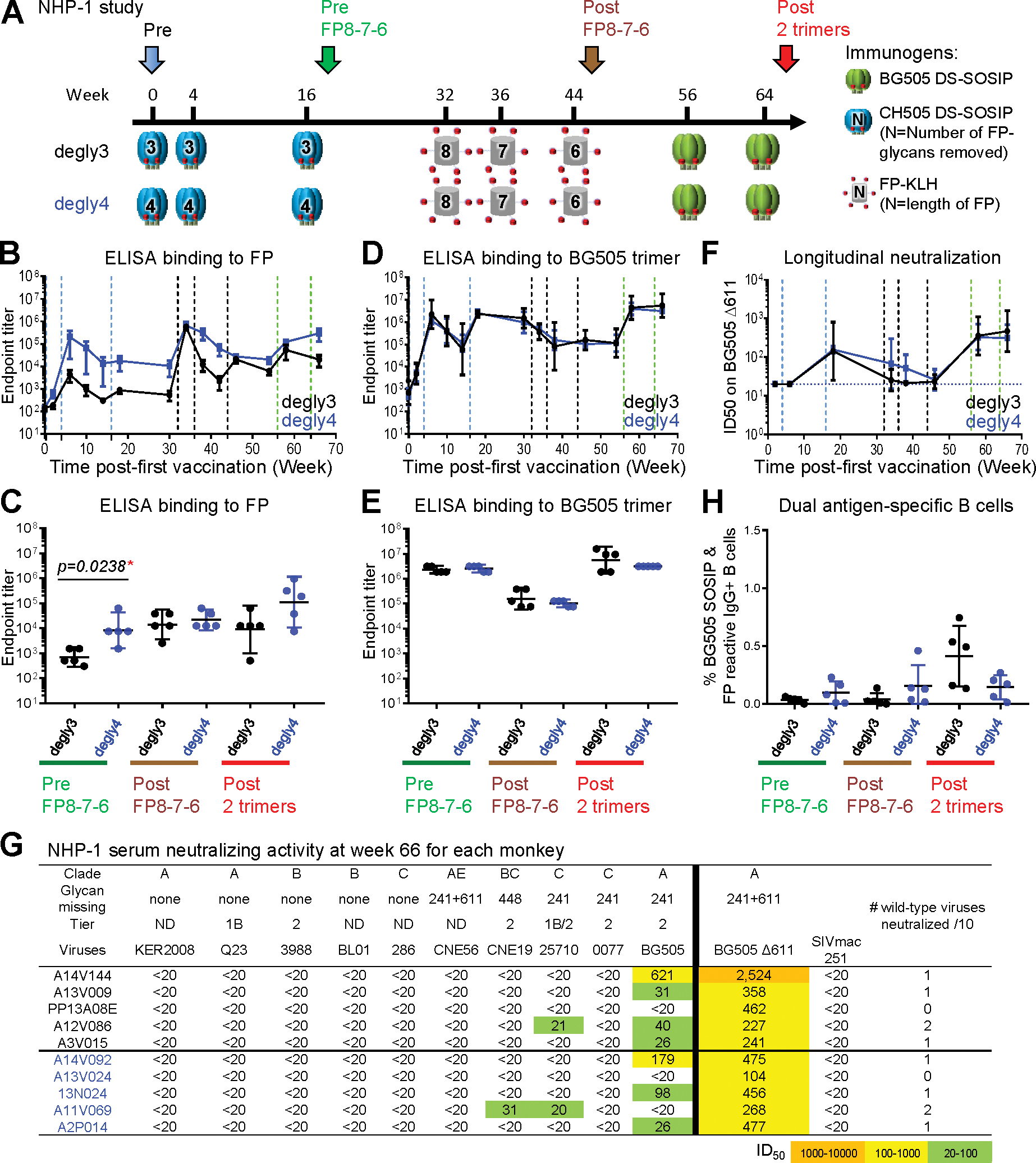
(A) Immunization schema. Two groups of animals were primed with three immunizations of CH505 degly3 or degly4, followed by a FP8–7-6-trimer-trimer boosting module. Key time points before immunization (Pre), before FP-KLH immunization (Pre FP8–7-6), after FP-KLH immunization (Post FP8–7-6), and the end of the study (Post two trimers) are indicated with different-colored arrows.
(B and D) ELISA IgG endpoint titers of serum antibodies binding to FP (B) or BG505 trimer (D). Vertical dashed lines represent immunizations.
(C and E) Comparison of serum antibody endpoint titers at three key time points to FP (C) or BG505 trimer (E).
(F) Comparison of the kinetics of the two groups on serum neutralization titers against BG505 Δ611 after the first immunization. Horizontal dashed line indicates minimum detection level.
(G) Neutralization ID50 titers on the 10-virus panel and BG505 Δ611 at week 66. Monkey ID is listed in the first column in black or blue letters representing the two groups. All FP8 sequences in the 10-strain panel were AVGIGAVF, the same as the immunogen, except 0077 (FP8:AVGIGAMF) and CNE56 (FP8:AVGIGAMI); this 10-strain panel was chosen as a sensitive means to detect neutralization by FP-targeting antibodies (Xu et al., 2018). ND, virus tier status has not been determined.
(H) Frequency of FP+/BG505+ B cells at three key time points as mean ± SD.
For all panels with error bars, geometric mean ± 95% CI are shown except where noted. p values calculated with Mann-Whitney two-tailed t test: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figures S2–S6 and Tables S1 and S2.
B cells with antigenic specificity for both FP and BG505 trimer probes (i.e., dual antigen-specific or double-positive B cells) were measured at three key time points, each 2 weeks after an immunization. These time points were pre FP8–7-6 (week 18), post-FP8–7-6 (week 46), and post 2 trimers (week 66) (Figure 2A). More than 0.5% of IgG+ B cells could bind to both antigens after BG505 trimer boosts in three animals of the degly3 group (Figure 2H). However, while FP and BG505 double-positive IgG+ B cells were detectable in the degly4 group at early time points, their frequency was not substantially increased at the end of the immunization scheme (Figure 2H, with both single- and double-positive B cells shown in Figure S1). In summary, the strategy of removing glycans proximal to FP from trimer immunogens succeeded in stimulating early ELISA responses to FP and Env trimer, but these responses were generally unable to neutralize wild-type HIV-1. We note that high-titer immune responses directed at sites of glycan removal have been observed for both CD4bs and glycan-V3 sites of vulnerability (Dubrovskaya et al., 2017; Escolano et al., 2019; Zhou et al., 2017); in prior cases, as observed here with FP, these responses could neutralize glycan-deleted, but not wild-type, viruses.
Dual Targeting of CD4bs and FP Yields Consistent CD4bs-Directed and Sporadic FP-Directed Responses (NHP-2 Study)
Elicitation of responses against multiple sites of vulnerability may be advantageous to an effective vaccine. Previously, we demonstrated that deletion of three glycans at N197, N276, and N462 from the CH505 trimer, which naturally lacks a fourth CD4bs-proximal glycan at N362, dramatically increases the immunogenicity of the CD4bs, with titers >104, though only against glycan-deleted viruses (Zhou et al., 2017). Because the CD4bs-glycan-deleted trimers did contain the FP site of vulnerability, we tested the CH505 Env trimer with four glycans around the CD4bs deleted (CD4bs-degly4) for its ability to induce both CD4bs and FP-directed responses.
Three groups of NHPs (groups A, B, and C) were immunized with the CD4bs-degly4 trimer, FP-KLH of various lengths, and the BG505 DS-SOSIP trimer (Figures 3A, 1A, and 1C). After two CD4bs-degly4 trimer primes, we observed high titers of anti-BG505 trimer responses but limited FP-directed responses in groups A and C at week 6 (Figures 3B–3E). Sera from both groups neutralized CH505 virus with four glycans removed around the CD4bs (CH505 CD4bs-degly4 virus) (Figure 3F) but did not neutralize wild-type CH505 virus at week 10 or 18 (Table S3). In contrast, group B, which received only one CD4bs-degly4 prime, did not show much neutralizing activity against the glycan-deleted or wild-type CH505 virus at week 10; however, another boost with CD4bs-degly4 at week 16 increased the neutralizing activity of group B against the glycan-deleted CH505 virus to a level similar to that of group A (Figure 3F). Overall, the neutralizing activity against wild-type CH505 and other heterologous viruses remained low (Table S3), as did the frequency of B cells double positive for both FP and Env trimer (Figure S1).
Figure 3. Dual Targeting of CD4bs and FP Yields Consistent CD4bs-Directed and Sporadic FP-Directed Neutralizing Responses.
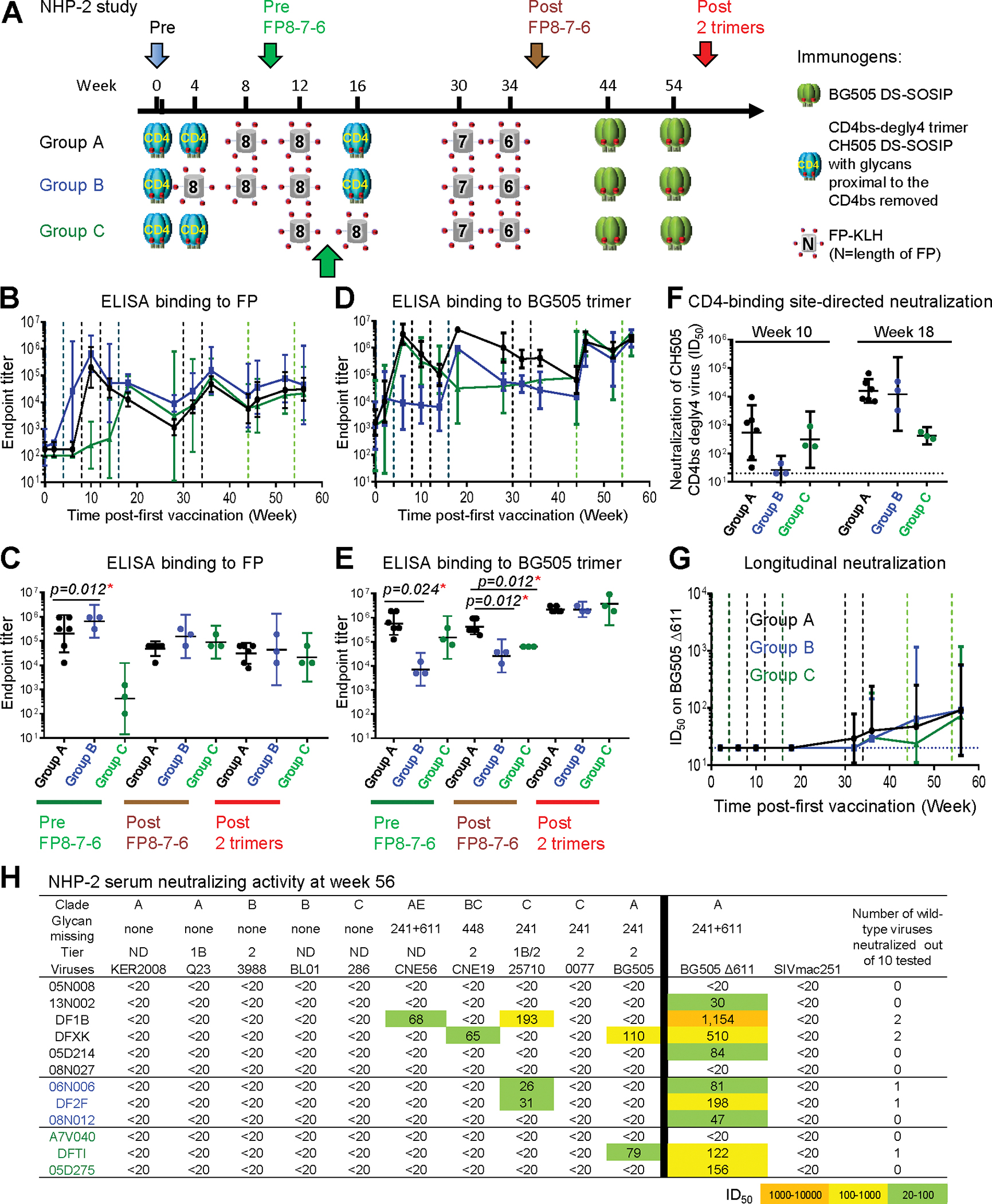
(A) Immunization schema. Key time points for analysis are indicated with colored arrows.
(B and D) ELISA IgG endpoint serum titers for binding FP (B) or BG505 trimer (D). Vertical dashed lines represent immunizations.
(C and E) Comparison of the three groups for serum endpoint titer to FP (C) or BG505 trimer (E) at three key time points.
(F) Neutralization ID50 titers of week 10 and 18 sera on CH505 CD4bs-degly4 virus. Horizontal dashed line indicates minimum detection level.
(G) Comparison of the three groups on serum neutralization against BG505 Δ611 over the course of the study.
(H) Neutralization ID50 titers on the 10-virus panel and BG505 D611 at the end of the study. This 10-strain panel was chosen as a sensitive means to detect neutralization by FP-targeting antibodies (Xu et al., 2018). Geometric mean ± 95% CI are shown. p values were calculated with Mann-Whitney two-tailed t test: *p < 0.05. ND, virus tier status has not been determined. See also Figures S2–S6 and Tables S1 and S3.
In an attempt to enhance FP-directed responses, immunizations using FP-KLH with different FP lengths followed by BG505 trimer were tested. In groups A and B, which received FP8 immunizations before week 10, high FP-binding responses were elicited at week 10 (pre FP8–7-6), and the titers did not increase after further boosts with FP8–7-6 (Figures 3B and 3C). Early anti-BG505 responses peaked after two CD4bs-degly4 immunizations in both groups A and C, and the response could be subsequently boosted with BG505 trimer, but not with FP-KLH. The group B regimen had lower peak anti-BG505 responses compared to the other two groups before BG505 boost. A third CH505 degly4 trimer boost at week 16 increased anti-BG505 response in group A to a higher level than groups B and C, and after the final two BG505 trimer immunizations (post 2 trimers), all groups reached similar late peak responses (Figures 3D and 3E).
In terms of neutralization, none of the animals developed serum responses capable of neutralizing the BG505 Δ611 mutant virus at week 18 (Figure 3G). At week 56 (post 2 trimers), neutralizing activity against BG505 Δ611 mutant virus was observed in several animals across groups (Figures 3G and 3H), but neutralizing activity against wild-type viruses was sporadic with only two animals showing measurable neutralization against BG505 (Figure 3H). Thus, CH505 CD4bs-degly4 trimers elicited immune responses to the CD4bs but failed to elicit substantial FP-directed responses, and boosting with FP-KLH or with BG505 trimer did not induce potent or broad neutralizing responses.
Priming with FP-KLH either Alone or in a Cocktail with Env Trimer Induces Cross-Clade Neutralizing Responses (NHP-3 Study)
FP-KLH priming and BG505 Env-trimer boost have been shown previously to elicit FP-directed responses in mice, guinea pigs, and NHPs (Xu et al., 2018). However, cocktails of FP-KLH with BG505 Env trimer have not been assessed for their ability to induce broadly neutralizing responses. Here, we compared the priming ability of a cocktail of FP-KLH and BG505 Env trimer versus FP-KLH alone in combination with our FP8–7-6-trimer-trimer boosting module (Figure 4A). Both groups elicited similar anti-FP IgG plasma responses (which effectively peaked after two immunizations), but the cocktail group elicited higher anti-BG505 ELISA responses at all time points (Figures 4B–4E). After two cocktail immunizations, the anti-BG505 ELISA titers rose to over 100,000, dropping at week 18 and rising at week 22 after the third immunization, after which the titers did not change dramatically, despite subsequent boosts. The FP-only primed group did not have high anti-BG505 Env-trimer responses at early time points, but responses increased after each of the final two trimer immunizations to reach titers similar to those of the cocktail-primed group by week 66.
Figure 4. Priming with FP-KLH, either Alone or in a Cocktail with Env Trimer, Induces Cross-Clade Neutralizing Responses.
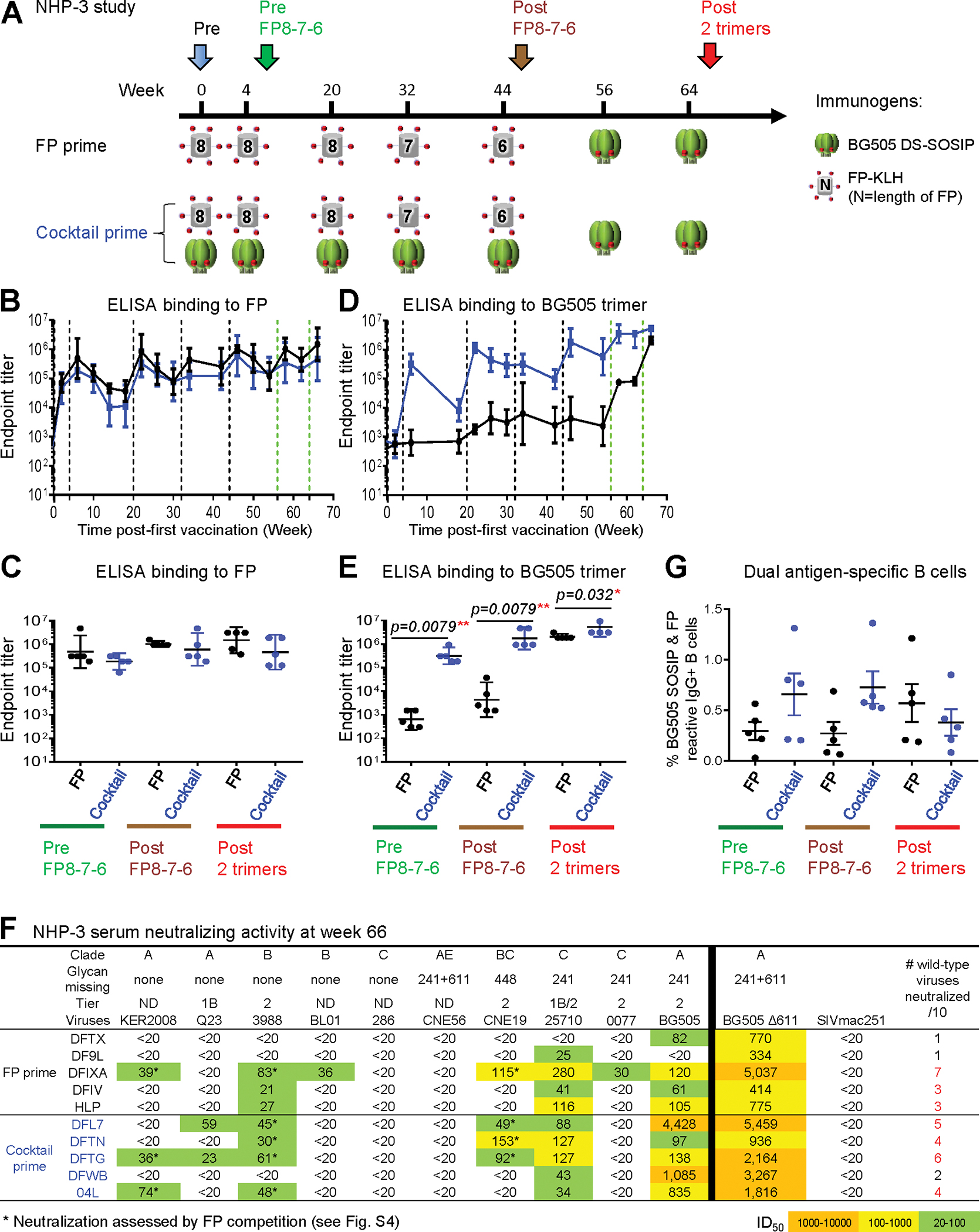
(A) Immunization schema.
(B and D) ELISA IgG endpoint serum titers for binding FP8 (B) or BG505 trimer (D). Vertical dashed lines represent immunizations.
(C and E) Comparison of the FP and cocktail groups for serum endpoint titer to FP (C) or BG505 trimer (E) at three key time points.
(F) Serum neutralization ID50 titers on the 10-virus panel and BG505 D611 at the end of the study in each monkey. This 10-strain panel was chosen as a sensitive means to detect neutralization by FP-targeting antibodies (Xu et al., 2018). ND, virus tier status has not been determined. In the last column, red numbers indicate serum neutralization of three or more wild-type viruses on the 10-strain panel. Asterisks indicates neutralization assessed by FP competition.
(G) Frequency of FP+/BG505+ B cells at three key time points.
Geometric mean ± 95% CI are shown, except in (G), which shows mean ± SD. p values were calculated with Mann-Whitney two-tailed t test: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. See also Figures S1–S6 and Tables S1, S4, and S5.
In terms of virus neutralization, the cocktail-primed group elicited significantly higher autologous neutralizing activity against wild-type BG505 virus with 6.7- and 13.4-fold higher ID50 titers at week 34 and week 46 (“post FP8–7-6”), respectively; by the end of the study (post 2 trimers), titers reached a geometric mean ID50 value of 560, which was 8.4-fold higher than the FP-only primed group (Figures 4F and S2; Table S4). In addition to higher autologous potency, the cocktail-primed animals also showed earlier neutralization breadth detected as early as week 22 after three immunizations, with titers continuing to increase after additional immunizations (Figure S2C). By week 66, sera from each of the 10 NHPs in the study neutralized at least one wild-type virus from the 10-strain panel (Figure 4F). One animal (DFIXA) in the FP-primed group neutralized 7 of 10 tested strains, and four animals (DFL7, DFTN, DFTG, and 04L) in the cocktail-primed group neutralized 4–6 viruses (Figure 4F). The 10-strain panel was chosen to provide a sensitive means to detect neutralization by FP-targeting antibodies (Xu et al., 2018), but high breadth on this panel (80%) may correspond to only 10%–20% breadth on the 208-strain panel; indeed, on the 12-strain global panel (deCamp et al., 2014), we observed only two strains to be neutralized by the week 66 NHP-3 sera (Figure S2D). FP competition of the plasma-neutralizing activities in each of these five animals confirmed the observed low-titer heterologous viral neutralization to be FP directed, with neutralizing activity on tier 2 viruses 3988, CNE19, and KER2008 significantly blocked by the presence of FP, but not by control peptide or medium (Figure S2E; Table S5).
To understand further the immune responses elicited by FP-only primed or by FP+trimer-cocktail-primed immunization, we analyzed the frequencies of antigen-specific B cells. We focused on double-positive B cells, reactive with both FP and Env trimer, as B cells encoding broadly neutralizing FP-directed antibodies that we previously identified (Kong et al., 2019) from vaccinated NHPs were double positive for FP and Env trimer. We found the cocktail-primed group to elicit a higher frequency of double-positive B cell responses at week 6 (pre FP8–7-6) and week 46 (post FP8–7-6) (Figures 4G and S1). The frequency of double-positive B cells in the cocktail-primed group decreased after two additional immunizations with trimer alone, so that by the end of the study, the frequency of double-positive B cells in the FP-only primed group was higher (Figures 4G and S1). This decrease could be due to repetitive immunizations with BG505; the last two boosts were the sixth and seventh immunizations with BG505 for the cocktail-primed group but were the first and second immunizations with BG505 for the FP-only primed group. Overall, priming with FP, either alone or in a cocktail with BG505 trimer, induced cross-clade FP-directed neutralizing responses. In addition to higher homologous neutralizing titers against the BG505 wild-type virus, the FP-trimer-cocktail-primed group elicited the earliest cross-clade responses (Figure S2C). Moreover, we note that the levels of double-positive B cells from the cocktail-primed group at week 6 (pre FP8–7-6) were remarkably high, ranging from 0.2% to 1.3% (Figure S3); this early elicitation of high-frequency double-positive B cells was substantially higher than observed in any of the NHP groups tested here (Figure S1) or in our previous study (Kong et al., 2019).
Env-Trimer Priming May Impede the Development of High-Titer FP-Directed Neutralizing Responses, but Env-Trimer Boosting Induces Breadth
Although the three NHP studies described above (NHP-1, NHP-2, and NHP-3) utilized different immunogens in various combinations, they shared a common FP8–7-6-trimer-trimer boosting module (Figure 5A). This enabled us to compare the impact of different primes. Notably, the FP-only or FP-Env-cocktail-primed groups yielded significantly higher neutralization on the 10-strain panel than groups that began with a trimer-only prime (p < 0.0001) (Figure 5B, left). We also observed FP-only or FP-Env-cocktail-primed groups to induce significantly higher ID50 titers against BG505 Δ611 than groups that began with a trimer-only prime (p = 0.0002) (Figure 5B, right).
Figure 5. Comparison of Different Vaccine Regimens that Use a Common Boosting Module Reveals FP or FP-Trimer Cocktail Priming to Imprint Broad FP-Directed HIV-Neutralizing Responses.
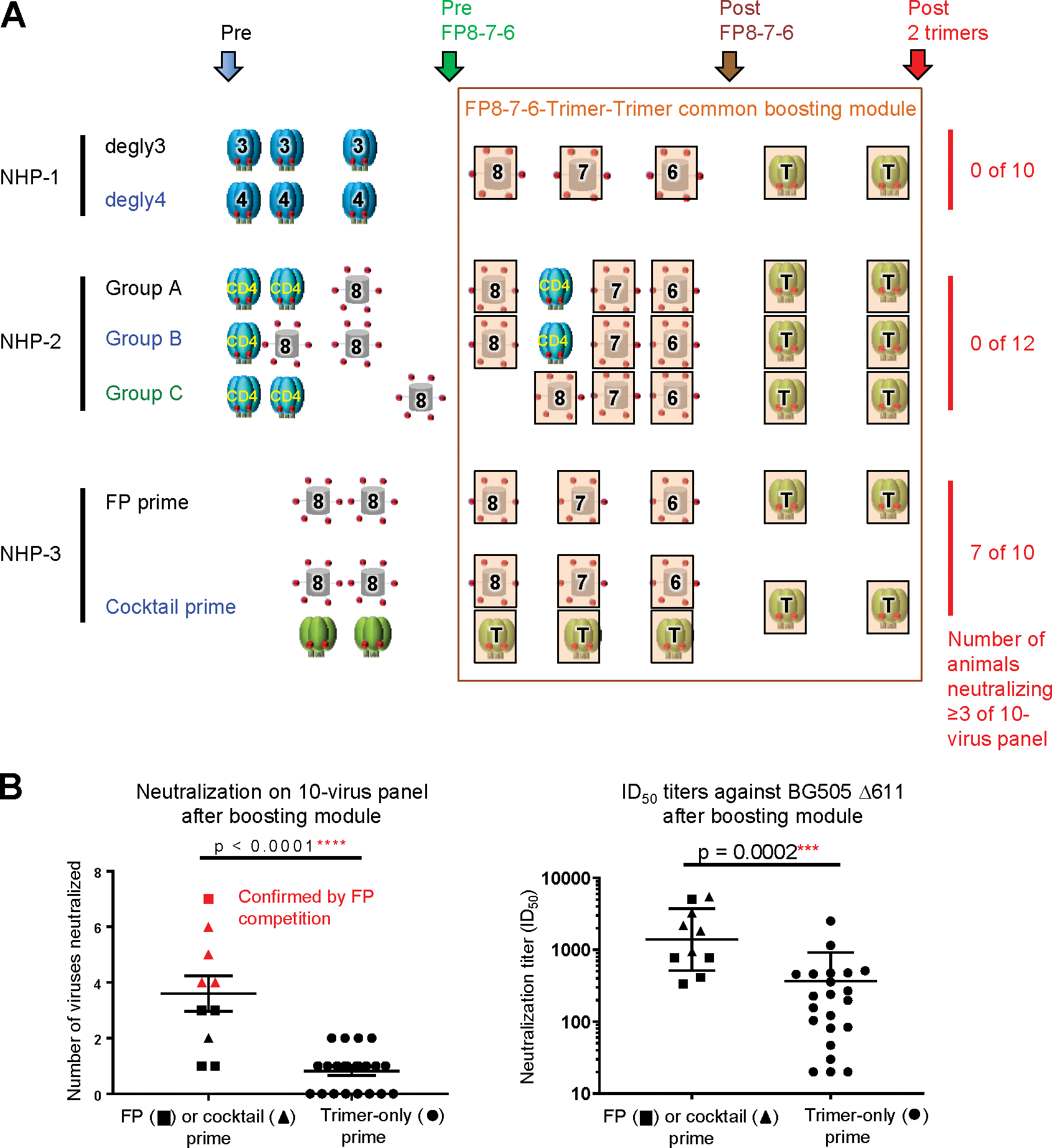
(A) Summary of the seven vaccine groups with shared key time points for analysis. Number of animals in each study with serum neutralization of at least three viruses on the 10-strain panel are listed on the right column. Key time points are indicated by colored arrows, and the common bosting module is outlined in brown.
(B) Comparison of neutralization breadth and titers after boosting module based on prime regimen. Neutralizing breadth at the end of the studies on the 10-virus panel and ID50 against BG505 D611 elicited with FP (squares) or cocktail (triangles) priming or trimer-only priming (circles) are shown. Mean ± SEM are shown for breadth, and geometric mean and SD are shown for ID50. p values were calculated with two-tailed, non-parametric Mann-Whitney t test. Points shown in red correspond to NHPs for which FP-directed neutralization has been confirmed by FP competition.
See also Figures S1–S3 and S6 and Tables S4 and S5.
We also analyzed the impact of the FP and trimer components of the boost module (Figure S4). Between pre FP8–7-6 and post FP8–7-6 time points, the geometric mean of BG505 Δ611 neutralization titers for all 32 NHPs rose from 38 to 57 ID50, but with no statistical significance. With respect to the separate NHP studies, the results were mixed, showing both increased (NHP-2 and NHP-3) and decreased (NHP-1) titers (Figure S4A). Between post FP8–7-6 and post 2 trimer time points, the geometric mean of BG505 Δ611 neutralization titers for all 32 NHPs rose from 57 to 327 ID50 (p < 0.0001) and showed a significant increase in each of the separate NHP studies (Figure S4B).
We further delineated the impact of each immunization in the boosting module (Figures S4C–S4G). Considering all 32 NHPs, none of the individual FP8–7-6 immunizations significantly impacted the BG505 Δ611 neutralization titers, while both of the trimer immunizations resulted in a significant increase (Figures S4C–S4G, leftmost panels). When segregated by individual study, four immunizations showed significant increases in ID50 titer; these were the FP8- and FP7-KLH immunizations for the NHP-3 study, the first trimer in the NHP-1 study, and the second trimer in the NHP-3 study (Figures S4C–S4G). The lack of statistical significance for many of the immunizations in the boosting module may relate to the overall low titer.
We also analyzed the impact of individual immunizations in individual groups (Figures S5A–S5D). Seven immunizations gave significant rise in BG505 Δ611 titer. These seven involved the penultimate trimer in NHP-1 degly3 group; the third degly4 trimer and the final trimer in the NHP-1 degly4 group; the penultimate and final trimers for NHP-3 FP-primed group; and the FP7-KLH+trimer and final trimer of the NHP-3-cocktail-primed group (Figures S5A–S5D). Overall, we observed diametrically opposed impacts of trimer immunization. With respect to boosting, all seven of the immunizations with statistically significant ID50 improvement involved trimer or FP+trimer; none involved FP-KLH alone (Figures S5A–S5D). In examining the boosting module, we also found that FP immunizations could raise or lower titers, but the trimer-trimer immunizations significantly raised titers overall, and in each of the separate studies (Figure S4). These observations are consistent with the prior finding that Env-trimer boost induces significant FP-directed neutralization (Xu et al., 2018). In contrast, with respect to priming, groups beginning with trimer-only immunization showed little neutralization on the 10-strain panel (Figure 5B, left). Thus, a single immunogen such as Env trimer can have completely different impacts on neutralization titer depending on context, which we infer to relate to the state of the humoral immune system at the time of immunization.
An Early Antigen-Specific B Cell Signature Correlates with FP-Directed Vaccine Outcome
To better understand the role of priming immunizations, we sought to identify immune parameters at the beginning of the boosting module that correlated with neutralizing responses. We analyzed a number of parameters, including plasma binding titers to FP and Env trimer (Table S1; Figures S6A–S6C), plasma neutralization titers against BG505 Δ611 (Figures 2F and 3G), and frequencies of antigen-specific B cells for FP, BG505, or both FP and BG505 (double-positive B cells) (Figures S1,S6D, and S6E).
For plasma binding titers to Env, significant correlations were observed between post FP8–7-6 titers and vaccine outcomes, but not between pre FP8–7-6 titers (prior to the boosting module) and vaccine outcomes (Figure S6). By contrast, strong correlation was observed between the number of wild-type viruses neutralized at the end of the study and the frequency of double-positive B cells before FP8–7-6 (Figure 6A); this correlation decreased post FP8–7-6 and was not significant at the end of the boosting module (Figure 6B). Contingency analysis indicated that for a desired breadth of at least 30% on the 10-strain panel at the end of the study, the frequency of double-positive B cells should be >0.35% before the boosting module (Figures 6C and 6D).
Figure 6. Identification of an Early B Cell Signature that Correlates with FP-Directed Vaccine Outcome.
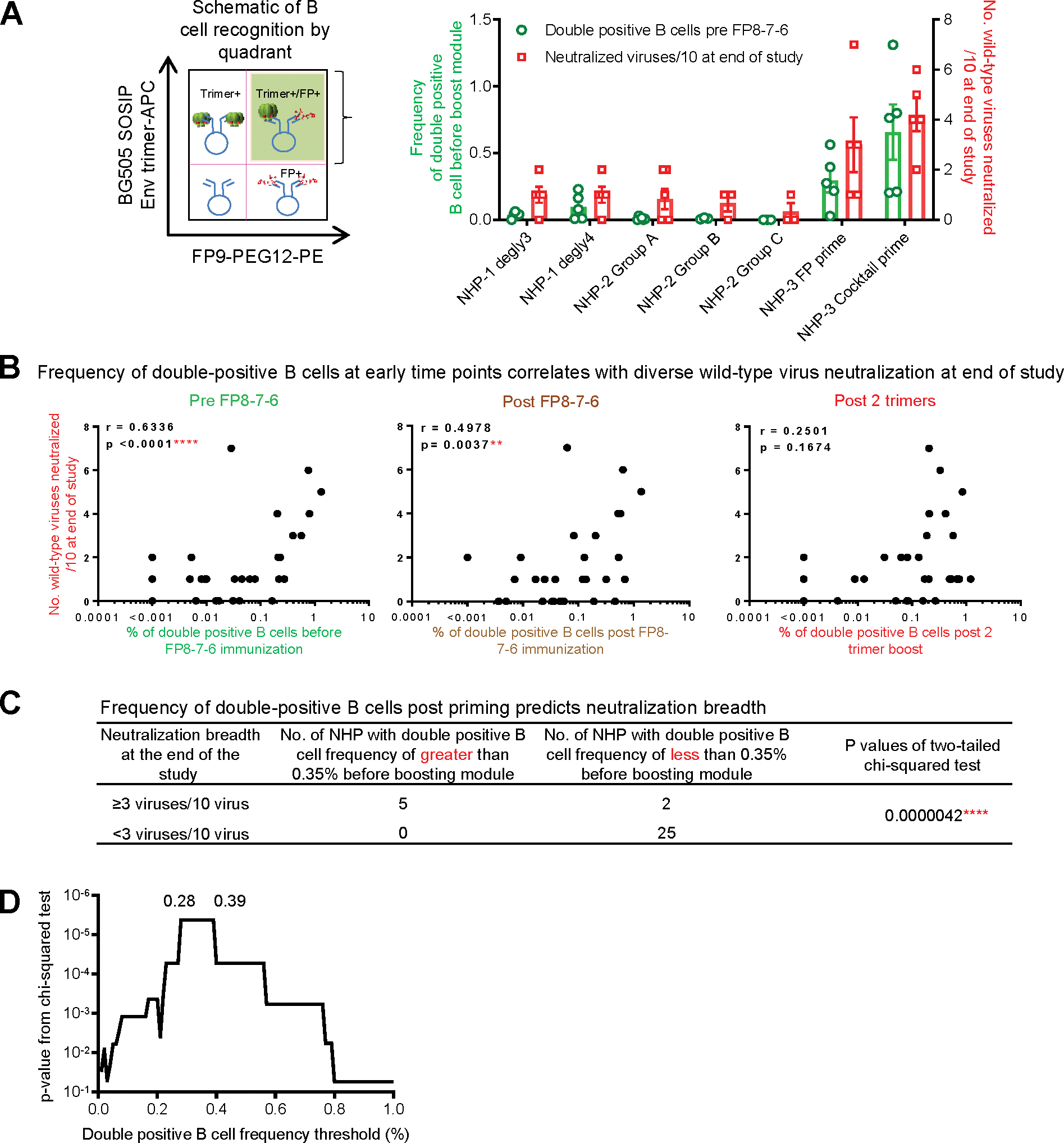
(A) Left: Schematic quadrant based on probe specific binding by IgG+ B cells to BG505 SOSIP and FP peptide highlighting the double-positive quadrant (shaded) corresponding to dual antigen-specific B cells. Right: Frequency of double-positive B cells at pre FP8–7-6 and neutralization breadth on the 10-virus panel at the end of the study from seven immunization regimens.
(B) Correlation of FP+/BG505+ double-positive B cells at pre FP8–7-6, post FP8–7-6, and post 2 trimer boost with neutralizing activity on the 10-virus panel at the end of the study.
(C) NHPs with frequency of FP+/BG505++ B cells >0.35% at pre FP8–7-6 are more likely to have a higher neutralization activity at the end of the study.
(D) Distribution of p value from (C) relative to frequency of double-positive B cells before FP8–7-6 immunization.
For (A), neutralization breadths and B cell frequencies were calculated as mean ± SEM. For (B), r and p values were calculated with two-tailed Pearson correlation analysis. For (C) and (D), p values were calculated with the chi-square test. See also Figures S3 and S6.
Overall, our analysis of immune system parameters for 32 NHPs revealed strong correlation between the frequency of double-positive B cells at the beginning of the boosting module and cross-reactive plasma neutralization at the end of the study approximately 1 year later. We were able to identify this correlation because our regimens utilized a common boosting module with different primes. In general, variation in regimens and in outcomes can be helpful in identifying underlying correlations. With respect to regimens, we note the boost module itself contained variations in intervals between different regimens; we analyzed the impact of these differing time intervals (Figures S5E–S5J) and found that longer time intervals tended to yield higher responses. With respect to outcomes, we note that our ability to identify correlates for improvement was dependent on the diverse effects induced by the different vaccine regimens tested, in which even regimens inducing weaker outcomes were critical to reveal correlates for improvement.
DISCUSSION
Elicitation of protective responses against HIV is likely to require complex immunization regimens that extend over many months (Andrabi et al., 2018; Bonsignori et al., 2017; Jardine et al., 2015; Kwong and Mascola, 2018). Currently, even for autologous-only neutralization, high-titer responses require many months to achieve with even the latest Env trimers and immunization schemes (Cirelli et al., 2019; Pauthner et al., 2017), with only the best of these schemes more recently assessed for its ability to protect against BG505 SHIV challenge (Pauthner et al., 2019). The current study seeks to use immune monitoring of developing B cells to understand how FP targeting might extend neutralization breadth from autologous-only to >50%. As elicited neutralization potency for FP-immunization is still suboptimal (Cheng et al., 2019; Kong et al., 2019; Xu et al., 2018), in this study, we relied on the neutralization fingerprint for broad FP-directed responses (Georgiev et al., 2013) and assessed breadth on a 10-strain panel, which is particularly sensitive to FP-directed neutralization (Xu et al., 2018). Even with this more sensitive panel, however, we could not detect much neutralization breadth with several of the immunization schemes (Figures 2 and 3); we could detect neutralization breadth only with FP or cocktail priming (Figure 4).
In natural HIV infection, broadly neutralizing plasma responses arise after years of infection (Hraber et al., 2014). Indeed, the extraordinary levels of somatic hypermutation observed with most known broadly neutralizing antibodies (Chuang et al., 2019; Scheid et al., 2009) may require long immunization regimens. There may therefore be a substantial advantage to understanding early B cell developmental parameters that are predictive of positive vaccine outcomes. Overall, our data indicate that an early antigen-specific B cell signature is significantly associated with the final plasma neutralization breadth (which was only achieved after extensive FP and trimer immunizations). The identified signature, based on the frequency of B cells capable of recognizing both Env and FP immunogens, appeared as early as week 6 and could potentially be used to prioritize vaccine immunogens and regimens based on their ability to achieve the identified double-positive signature.
We note that in a separate study (Kong et al., 2019), where we identify and track five broadly neutralizing FP-directed lineages in NHP, all of the broadly neutralizing antibodies derive from B cells with this double-positive signature. However, with naturally occurring FP-directed antibodies, the presence of the signature may not be universal, as PGT151 (Blattner et al., 2014) and ACS202 (van Gils et al., 2016; Yuan et al., 2019) have lower affinities for FP than the murine or NHP antibodies induced by FP and Env vaccination (Kong et al., 2019; Xu et al., 2018). The double-positive signature we identified for vaccine-induced neutralization breadth may thus be FP vaccine specific and may not relate to FP-directed antibodies from natural infection.
We also looked for correlations between the frequency of single-positive (FP+ or BG505+) B cells and neutralization breadth at the end of the study and observed weak correlation (r = 0.3898, p = 0.0274) between single BG505-positive B cells (BG505+FP−) at the post FP8–7-6 time point and endpoint breadth (Figures S5E–S5J). This correlation likely relates to the fact that, at this point in the boosting module, only FP immunogens and not Env-trimer immunogens have been administered, and thus, BG505-positive responses are indicative of an FP-induced cross-reactive response to Env trimer.
Our results raise multiple questions. Why does priming with Env trimer alone fail? Why is the FP+trimer cocktail so much better? How can the frequency of double-positive B cells be increased? The last question likely has numerous answers, but we believe one will be to utilize approaches involving cocktails containing both Env and FP. While a cocktail may have parameters such as ratio of components, it can still be embodied as a single vaccine product and therefore requires fewer variables to optimize than a prime-boost regimen. Regardless of the immunogen or regimen, the early B cell signature identified here is likely to play a role in obtaining highly protective broadly neutralizing titers from FP vaccination.
In immunological imprinting, an early immune experience shapes later immunological outcomes (Davenport et al., 1953; Francis, 1960; Kouyos et al., 2018; Lessler et al., 2012; Yu et al., 2008). While most prior imprinting studies relate to natural infection, not the vaccination regimens examined in the current study, we nonetheless observed a similar immunological trend. Specifically, it appears that unless FP immunogens are simultaneously introduced with trimer during the initial priming exposure, trimer priming during the very first exposure to HIV-1 immunogens acts to narrow the breadth of later immune responses. We speculate that this may reflect “antigenic imprinting,” whereby humoral responses to initial, trimer-only priming lay a foundation of memory B cells directed primarily against non-FP epitopes, which then tend to outcompete FP-binding B cells during germinal center formation upon subsequent exposure to Env trimers. If correct, this B cell imprinting mechanism would resemble the impact of initial childhood exposure to influenza A virus hemagglutinin antigens in determining outcomes to subsequent exposures (Gostic et al., 2016; Worobey et al., 2014). A potential implication in the general vaccine strategy would be to use as a priming immunogen the most conserved region of the most neutralizing epitope in order to achieve a broadly neutralizing vaccine outcome.
Our early B cell signature appears to identify an essential characteristic imprinted by the priming immunogen to achieve a final outcome of broad neutralization. Our analysis is similar in some respects to immune correlate studies (Tomaras and Plotkin, 2017), and there are likely to be multiple immunological factors that contribute to imprinting. In addition to the B cell mechanism described above, T cell mechanisms are likely to play a role. For example, as shown with vaccination for yellow fever, the initial immunization imprints a lifelong dominant response, epigenetically encoded in antigen-specific T cells (Akondy et al., 2017). It seems likely that the robust T cell help provided by KLH induced a more substantial and more durable response than the T cell help provided by HIV Env. We note that the similarities of the responses observed for FP-only and FP+Env-trimer immunizations may be more consistent with T-cell-based mechanisms of imprinting. Specifically, the presence of Env trimer in the prime would not be expected to diminish the T cell response to KLH but would be expected to introduce B cell competition and thereby diminish FP-directed responses. Further experiments measuring longitudinal T cell responses will be needed to delineate a complete mechanistic basis for the imprinting observed here with FP vaccination to elicit broad HIV-neutralizing responses.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Peter D. Kwong (pdkwong@nih.gov).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
NHP studies
Animals were housed and cared for in accordance with local, state, federal, and institute policies in an American Association for Accreditation of Laboratory Animal Care-accredited facility (Bioqual Inc, MD). All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Vaccine Research Center, NIAID, NIH, under protocols VRC #16–667.1 for the NHP-1 study, #16–666.1 for the NHP-2 study, and #16–667.2 for the NHP-3 study. Female or male Indian rhesus macaques used in the studies were 2–14 years old and had body weights of 4–10 kg. Animals were evenly distributed to different groups in each of the studies based on body weight. All incoming animals were healthy (B-virus, SIV, SRV, and STLV negative) and without previous exposure to HIV or SHIV; they were not involved in previous procedures and were drug tested naive. For each immunization, a total of 100 μg of specified, filter-sterilized immunogen (in case of cocktail, two immunogens were mixed at 50 μg each) in PBS and 200 μL of Adjuplex (Sigma-Aldrich Inc, MO or Adjuplex equivalent formulated based on US Patent 6,676,958 B2) were mixed in a total volume of 1 mL and injected via a needle syringe to the caudle thigh of the two hind legs at 500 μL each. Whole blood was collected for serological analyses. Plasma and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll density gradient centrifugation.
Cell Lines
FreeStyle 293-F cells were from ThermoFisher Scientific Inc (cat# R79007). Cells were maintained in FreeStyle 293 Expression Medium. The cell line was used directly from the commercial sources and cultured following manufacturer suggestions as described in Method Details below.
METHOD DETAILS
FP-KLH immunogens
FP-KLH immunogens were prepared as described previously (Kong et al., 2019; Xu et al., 2018). Three HIV-1 fusion peptides were synthesized (GenScript) with a free N-terminal amine group and a Cys appended at the C terminus, including FP8 (AVGIGAVF), FP7 (AVGIGAV) and FP6 (AVGIGA). The carrier protein, KLH (Thermo-Scientific), was activated with m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS, Sigma) and then ligated to the Cys thiol group of the FP peptides to make FP-KLH conjugates. The conjugates were verified antigenically with FP specific antibodies, VRC34.01, PGT151 and ACS202.
HIV-1 envelope trimer immunogens and probes
Glycan-deleted CH505 Env trimers were prepared in chimeric format with gp41 and the N and C termini of gp120 from BG505 DS-SOSIP, and mutations to remove the selected glycan sites, as described previously (Zhou et al., 2017). Non-tagged or Avi-tagged Env trimers were generally produced in transiently transfected 293F cells as previously described (Pancera et al., 2014; Sanders et al., 2013). The trimer proteins were purified from the cell culture supernatant by 2G12 or VRC01 affinity chromatography, followed by gel filtration with a Superdex200 16/60HL column and then negative selection to remove V3-exposed trimers with a 447–52D affinity column. The antigenicity of the trimers was determined using the Meso Scale Discovery (MSD) platform as previously described (Kwon et al., 2015). A few of the trimer immunogens (NHP-1, week 64 and NHP-3, weeks 56 and 64) were produced in stable CHO cell lines and purified using non-affinity chromatography; the antigenicity of the 293F- and CHO-produced trimers, however, was virtually identical as assessed by MSD. To prepare sorting probes, the Avi-tagged trimers were biotinylated using the BIRA500 kit (Avidity, LLC) and purified by gel filtration chromatography, and the biotinylated trimers were coupled to Streptavidin-APC (Life Technologies).
FP probes
To prepare the FP probes, linear peptide comprising the N-terminal 8 or 9 residues of FP (FP8, as above; FP9, AVGIGAVFL) were synthesized and attached to biotin through a polyethylene glycol linker (FP-PEG-biotin ordered from Genscript). To prepare the FP9-PEG12-PE sorting probe, FP9-PEG12-biotin was ligated to streptavidin-PE (Invitrogen).
Negative-stain electron microscopy
Samples were diluted with a buffer containing 10 mM HEPES, pH 7.0, and 150 mM NaCl and adsorbed to a freshly glow-discharged carbon-film grid. The grid was washed with the same buffer, and proteins were stained with 0.7% uranyl formate. An FEI Tecnai T20 electron microscope equipped with a 2k × 2k Eagle CCD camera and operated at 200 kV was used to collect negative-stain datasets. Micrographs were collected semi-automatically using SerialEM (Mastronarde, 2005) at a magnification of 100,000 × corresponding to a pixel size of 0.22 nm. 2D classification was performed using EMAN2 (Tang et al., 2007).
Enzyme-linked immunosorbent assay (ELISA)
Animal sera were assessed for binding to FP using FP8-PEG-biotin as previously described (Cheng et al., 2019). Streptavidin coated plates (Thermoscientific, Rockford, IL) were coated overnight at 4°C with FP8-PEG-biotin. Sera were heat-inactivated at 56°C for 1 hour and assessed at 7-point 5-fold dilutions starting at 1:100 in B3T buffer (150 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 3.3% fetal bovine serum, 2% bovine albumin, 0.07% Tween 20, 0.02% thimerosal). Goat anti-NHP IgG antibody conjugated to horseradish peroxidase (KPL, Gaithersburg, MD) diluted 1:5,000 in B3T buffer was added. Plates were developed with tetramethylbenzidine (TMB) substrate (SureBlue; KPL, Gaithersburg, MD) for 10 min before the addition of 1 N sulfuric acid (Fisher Chemical, Fair Lawn, NJ) to stop the reaction and read at 450 nm (SpectraMax using SoftMax Pro, version 5, software; Molecular Devices, Sunnyvale, CA). ELISA against BG505 DS-SOSIP.664 was performed with a modified procedure based on previously reported method with lectin captured trimer (Georgiev et al., 2015). Ninety-six-well plates (Costar High Binding Half-Area; Corning, Kennebunk, ME) were coated overnight at 4°C with 2 μg/ml snowdrop lectin from Galanthus nivalis (Sigma-Aldrich, St. Louis, MO) in PBS. Plates were washed 5 times with PBS plus 0.05% Tween-20 and then were blocked with 5% skim milk in PBS for 60 min at room temperature, followed by trimer capture with 2 μg/ml BG505 DS-SOSIP.664 in 10% FBS-PBS for 2 hours at room temperature. Next, 7-point serially diluted (5-fold; starting dilution 1:100) monkey plasma in 0.2% Tween-PBS buffer was added and incubated for 1 hour at room temperature. Afterward, goat anti-NHP IgG antibody conjugated to horseradish peroxidase (Alpha Diagnostic International, San Antonio, TX) diluted 1:5,000 in 0.2% Tween-PBS buffer was added at 50 μl/well and incubated for 60 min. Plates were washed five times with PBS plus 0.05% Tween-20 and developed with 50 μl/well tetramethylbenzidine (TMB) (SureBlue; KPL, Gaithersburg, MD) for 10 min. The reactions were stopped by addition of 50 μl/well 1 N sulfuric acid (Fisher Chemical, Fair Lawn, NJ). Plates were read at 450 nm (SpectraMax using SoftMax Pro, version 5, software; Molecular Devices, Sunnyvale, CA), and the optical densities (OD) were subtracted for the nonspecific horseradish peroxidase background activity. The endpoint titer was defined as the reciprocal of the greatest dilution with an OD value above 0.1 (2 times average raw plate background).
Neutralization assays
Serum neutralization was assessed with a single round virus infection assay using TZM-bl target cells as previously described (Kong et al., 2016). Briefly, HIV-1 Env-pseudotyped virus stocks were generated by cotransfecting 293T cells with an Env expression plasmid and a pSG3ΔEnv backbone. Animal sera were heat-inactivated at 56°C for 1 hour and assessed at 8-point 4-fold dilutions starting at 1:20. Virus stocks and sera were mixed in a total volume of 50 μL and incubated at 37°C for 1 hr. TZM-bl cells (20 μl, 0.5 million/ml) were then added to the mixture and incubated at 37°C. On day 2, 130 μL cDMEM was added to feed the cells. On day 3, cells were lysed and assessed for luciferase activity (RLU). Data were fit to a 5-parameter hill slope equation by nonlinear regression, and the 50% inhibitory dilutions (ID50) were determined.
Neutralization with peptide competition was performed as described previously (Kong et al., 2016; Xu et al., 2018). Briefly, NHP plasma was tested at a single-point dilution that resulted in 50%–80% neutralization of the virus. 10 μL of plasma was mixed with 5 μL of control media, PEGylated FP9, or PEGylated non-cognate FLAG peptide, and the mixture incubated at 37°C for 30 minutes; 35 μL of each virus was then added, and incubation at 37°C continued for 30 minutes. The final peptide concentration was 12.5 pM. TZM-bl cells were added, incubated, fed, and lysed, and luciferase activity assessed, as described above. The assay was performed in duplicated wells and repeated at least three times.
FACS analysis of PBMC
FACS analysis of PBMC was performed as described previously (Kong et al., 2019). Briefly, NHP PBMCs were stained with LIVE/DEAD fixable violet dead cell stain (Life Technologies), washed, and then stained with a cocktail of anti-human antibodies, including CD3 (clone SP34–2; BD Biosciences), CD4 (clone OKT4; BioLegend), CD8 (clone RPA-T8; BioLegend), CD14 (clone M5E2; BioLegend), CD20 (clone 2H7; BioLegend), IgG (G18–145; BD Biosciences), and IgD (Dako, polyclonal), and with fluorescently labeled trimer probe (BG505 DS-SOSIP) for 15 mins, and followed by peptide probe FP9-PEG12-PE for another 15 mins. Stained PBMCs were analyzed on BD LSRFortessa X-50. Vivid−CD3−CD4−CD8−CD14−CD20+IgG+IgD− memory B cells that were positively stained with both trimer and peptide probes were considered FP and trimer dual-specific memory B cells. The analysis of the PBMCs was performed using FlowJo.
QUANTIFICATION AND STATISTICAL ANALYSIS
The statistical difference of neutralization titers or breadth, ELISA titers or B cell frequency between different groups was determined by performing un-paired non-parametric two-tailed Mann-Whitney tests. Two-tailed Pearson correlation coefficients with 95% confidence intervals were used to calculate the correlation between frequency of double-positive B cells, ELISA endpoint titers, and neutralization potency and breadth. Two-tailed chi-square tests were used to determine the predictability of obtaining double-positive B cells frequencies with neutralization breadth at the end of study. Two-tailed non-parametric Wilcoxon matched pairs signed rank tests were performed to compare Δ611 neutralization titers between immunizations in the FP-8–7-6-trimer-trimer boosting sequence. The statistical analyses were performed using GraphPad Prism 7. A p value of 0.05 or lower was considered as statistically significant.
Supplementary Material
Highlights.
Immunization in 32 rhesus macaques reveals FP priming to imprint cross-clade responses
Identifying an early B cell signature predictive of vaccine outcome
Priming with a cocktail of FP and trimer elicits the earliest neutralizing responses
B cell immune monitoring enables vaccine insights, even without serum neutralization
ACKNOWLEDGMENTS
We thank J. Hill, E. Smit, R. Nguyen, D. Ambrozak, and S.P. Perfetto of the Flow Cytometry Core at the Vaccine Research Center (VRC) for assistance with B cell sorting; J. Noor and E. McCarthy of the Translational Research Program and Bioqual veterinary technical staff; K. Saunders and B.F. Haynes for discussions on CH505; J. Stuckey for assistance with figures; members of the Non-human Primate Immunogenicity Core at the VRC for assistance with the NHP studies; and members of the Virology Laboratory and Vector Core, VRC, for discussions and comments on the manuscript. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), by federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001, and by the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium. M.W. was supported by a grant from the David and Lucile Packard Foundation.
CONSORTIA
The VRC Production Program includes Nadia Amharref, Nathan Barefoot, Christopher Barry, Elizabeth Carey, Ria Caringal, Kevin Carlton, Naga Chalamalsetty, Adam Charlton, Rajoshi Chaudhuri, Mingzhong Chen, Peifeng Chen, Nicole Cibelli, Jonathan W. Cooper, Hussain Dahodwala, Marianna Fleischman, Julia C. Frederick, Haley Fuller, Jason Gall, Isaac Godfroy, Daniel Gowetski, Krishana Gulla, Vera Ivleva, Lisa Kueltzo, Q. Paula Lei, Yile Li, Venkata Mangalampalli, Sarah O’Connell, Aakash Patel, Erwin Rosales-Zavala, Elizabeth Scheideman, Nicole A. Schneck, Zachary Schneiderman, Andrew Shaddeau, William Shadrick, Alison Vinitsky, Sara Witter, Yanhong Yang, and Yaqiu Zhang.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107981.
REFERENCES
- Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. (2017). Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrabi R, Bhiman JN, and Burton DR (2018). Strategies for a multi-stage neutralizing antibody-based HIV vaccine. Curr. Opin. Immunol. 53, 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, de la Peña AT, Cupo A, Julien JP, van Gils M, Lee PS, et al. (2014). Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity 40, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonsignori M, Liao HX, Gao F, Williams WB, Alam SM, Montefiori DC, and Haynes BF (2017). Antibody-virus co-evolution in HIV infection: paths for HIV vaccine development. Immunol. Rev. 275, 145–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR (2002). Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2, 706–713. [DOI] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, and Wyatt RT (2004). HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5, 233–236. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. (1994). Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027. [DOI] [PubMed] [Google Scholar]
- Cheng C, Xu K, Kong R, Chuang GY, Corrigan AR, Geng H, Hill KR, Jafari AJ, O’Dell S, Ou L, et al. (2019). Consistent elicitation of cross-clade HIV-neutralizing responses achieved in guinea pigs after fusion peptide priming by repetitive envelope trimer boosting. PLoS ONE 14, e0215163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Zhou J, Acharya P, Rawi R, Shen CH, Sheng Z, Zhang B, Zhou T, Bailer RT, Dandey VP, et al. (2019). Structural Survey of Broadly Neutralizing Antibodies Targeting the HIV-1 Env Trimer Delineates Epitope Categories and Characteristics of Recognition. Structure 27, 196–206.e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli KM, Carnathan DG, Nogal B, Martin JT, Rodriguez OL, Upadhyay AA, Enemuo CA, Gebru EH, Choe Y, Viviano F, et al. (2019). Slow delivery immunization enhances HIV neutralizing antibody and germinal center responses via modulation of immunodominance. Cell 177, 1153–1171.e1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe JE Jr. (2016). Teaching a clone to walk, one step at a time. Cell 166, 1360–1361. [DOI] [PubMed] [Google Scholar]
- Davenport FM, Hennessy AV, and Francis T Jr. (1953). Epidemiologic and immunologic significance of age distribution of antibody to antigenic variants of influenza virus. J. Exp. Med. 98, 641–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, et al. (2014). Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 88, 2489–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Bhiman JN, Roark RS, Schramm CA, Gorman J, Chuang GY, Pancera M, Cale EM, Ernandes MJ, Louder MK, et al. (2016). New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J. Virol. 90, 76–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. (2012). Highly conserved protective epitopes on influenza B viruses. Science 337, 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovskaya V, Guenaga J, de Val N, Wilson R, Feng Y, Movsesyan A, Karlsson Hedestam GB, Ward AB, and Wyatt RT (2017). Targeted N-glycan deletion at the receptor-binding site retains HIV Env NFL trimer integrity and accelerates the elicited antibody response. PLoS Pathog. 13, e1006614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbelding EJ, Post DJ, Stemmy EJ, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, and Fauci AS (2018). A universal influenza vaccine: the strategic plan for the national institute of allergy and infectious diseases. J. Infect. Dis. 218, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A, Gristick HB, Abernathy ME, Merkenschlager J, Gautam R, Oliveira TY, Pai J, West AP Jr., Barnes CO, Cohen AA, et al. (2019). Immunization expands B cells specific to HIV-1 V3 glycan in mice and macaques. Nature 570, 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis T Jr. (1960). On the doctrine of original antigenic sin. Proc. Am. Phil. Soc. 104, 572–578. [Google Scholar]
- Georgiev IS, Doria-Rose NA, Zhou T, Kwon YD, Staupe RP, Moquin S, Chuang GY, Louder MK, Schmidt SD, Altae-Tran HR, et al. (2013). Delineating antibody recognition in polyclonal sera from patterns of HIV-1 isolate neutralization. Science 340, 751–756. [DOI] [PubMed] [Google Scholar]
- Georgiev IS, Joyce MG, Yang Y, Sastry M, Zhang B, Baxa U, Chen RE, Druz A, Lees CR, Narpala S, et al. (2015). Single-chain soluble BG505.SOSIP gp140 trimers as structural and antigenic mimics of mature closed HIV-1 Env. J. Virol. 89, 5318–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, and Zolla-Pazner S (1992). Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66, 7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostic KM, Ambrose M, Worobey M, and Lloyd-Smith JO (2016). Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, and Korber BT (2014). Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS 28, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kang BH, Pancera M, Lee JH, Tong T, Feng Y, Imamichi H, Georgiev IS, Chuang GY, Druz A, et al. (2014). Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature 515, 138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine JG, Ota T, Sok D, Pauthner M, Kulp DW, Kalyuzhniy O, Skog PD, Thinnes TC, Bhullar D, Briney B, et al. (2015). HIV-1 Vaccines. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Burke V, Totrov M, Williams C, Cardozo T, Gorny MK, Zolla-Pazner S, and Kong XP (2010). Conserved structural elements in the V3 crown of HIV-1 gp120. Nat. Struct. Mol. Biol. 17, 955–961. [DOI] [PubMed] [Google Scholar]
- Kong R, Xu K, Zhou T, Acharya P, Lemmin T, Liu K, Ozorowski G, Soto C, Taft JD, Bailer RT, et al. (2016). Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong R, Duan H, Sheng Z, Xu K, Acharya P, Chen X, Cheng C, Dingens AS, Gorman J, Sastry M, et al. (2019). Antibody lineages with vaccine-induced antigen-binding hotspots develop broad HIV neutralization. Cell 178, 567–584 e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos RD, Rusert P, Kadelka C, Huber M, Marzel A, Ebner H, Schanz M, Liechti T, Friedrich N, Braun DL, et al. ; Swiss HIV Cohort Study (2018). Tracing HIV-1 strains that imprint broadly neutralizing antibody responses. Nature 561, 406–410. [DOI] [PubMed] [Google Scholar]
- Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, and Mascola JR (2018). HIV-1 vaccines based on antibody identification, B cell ontogeny, and epitope structure. Immunity 48, 855–871. [DOI] [PubMed] [Google Scholar]
- Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessler J, Riley S, Read JM, Wang S, Zhu H, Smith GJ, Guan Y, Jiang CQ, and Cummings DA (2012). Evidence for antigenic seniority in influenza A (H3N2) antibody responses in southern China. PLoS Pathog. 8, e1002802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HX, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. ; NISC Comparative Sequencing Program (2013). Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature 496, 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. [DOI] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Leung S, Graepel K, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, et al. (2013). Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibod. Science 340, 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088.e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-induced protection from homologous tier 2 SHIV challenge in nonhuman primates depends on serum-neutralizing antibody titers. Immunity 50, 241–252.e246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner MR, Cavacini LA, Emes CL, Power J, and Byrn R (1993). Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune. Defic. Syndr. 6, 7–14. [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Peña AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders KO, Verkoczy LK, Jiang C, Zhang J, Parks R, Chen H, Housman M, Bouton-Verville H, Shen X, Trama AM, et al. (2017). Vaccine induction of heterologous tier 2 HIV-1 neutralizing antibodies in animal models. Cell Rep. 21, 3681–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. (2009). Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640. [DOI] [PubMed] [Google Scholar]
- Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. (2016). Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell 165, 813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46. [DOI] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, and Sodroski J (1993). Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67, 3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaras GD, and Plotkin SA (2017). Complex immune correlates of protection in HIV-1 vaccine efficacy trials. Immunol. Rev. 275, 245–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, and Katinger H (1996). Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gils MJ, van den Kerkhof TL, Ozorowski G, Cottrell CA, Sok D, Pauthner M, Pallesen J, de Val N, Yasmeen A, de Taeye SW, et al. (2016). An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat. Microbiol. 2, 16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. (2003). Antibody neutralization and escape by HIV-1. Nature 422, 307–312. [DOI] [PubMed] [Google Scholar]
- Worobey M, Han GZ, and Rambaut A (2014). Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc. Natl. Acad. Sci. USA 111, 8107–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Acharya P, Kong R, Cheng C, Chuang GY, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M, et al. (2018). Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat. Med. 24, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, et al. (2008). Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Cottrell CA, Ozorowski G, van Gils MJ, Kumar S, Wu NC, Sarkar A, Torres JL, de Val N, Copps J, et al. (2019). Conformational plasticity in the HIV-1 fusion peptide facilitates recognition by broadly neutralizing antibodies. Cell Host Microbe 25, 873–883.e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Doria-Rose NA, Cheng C, Stewart-Jones GBE, Chuang GY, Chambers M, Druz A, Geng H, McKee K, Kwon YD, et al. (2017). Quantification of the impact of the HIV-1-glycan shield on antibody elicitation. Cell Rep. 19, 719–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Lynch RM, Chen L, Acharya P, Wu X, Doria-Rose NA, Joyce MG, Lingwood D, Soto C, Bailer RT, et al. (2015). Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.


