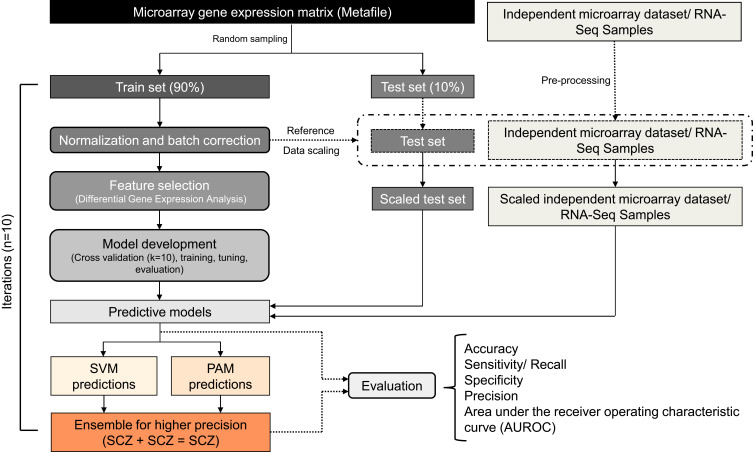
Figure 1.
The workflow depicts the steps involved in sample processing and class prediction analysis. A raw meta-file with common genes (rows) across microarray datasets and samples (columns) was used for the analysis. Samples from the raw meta-file were shuffled and subjected to a random selection of train and test data (9:1) generating ten iterations each. Each iteration of train data was independently quantile normalized. The preprocessed train data was used for feature selection and model training. Normalization and batch correction for test data, independent dataset, and RNA-Seq data were performed using train data as a reference. The performance of the individual models was compared with the ensemble learning. The models were also evaluated for their cross-platform compatibility using RNA-Seq samples.