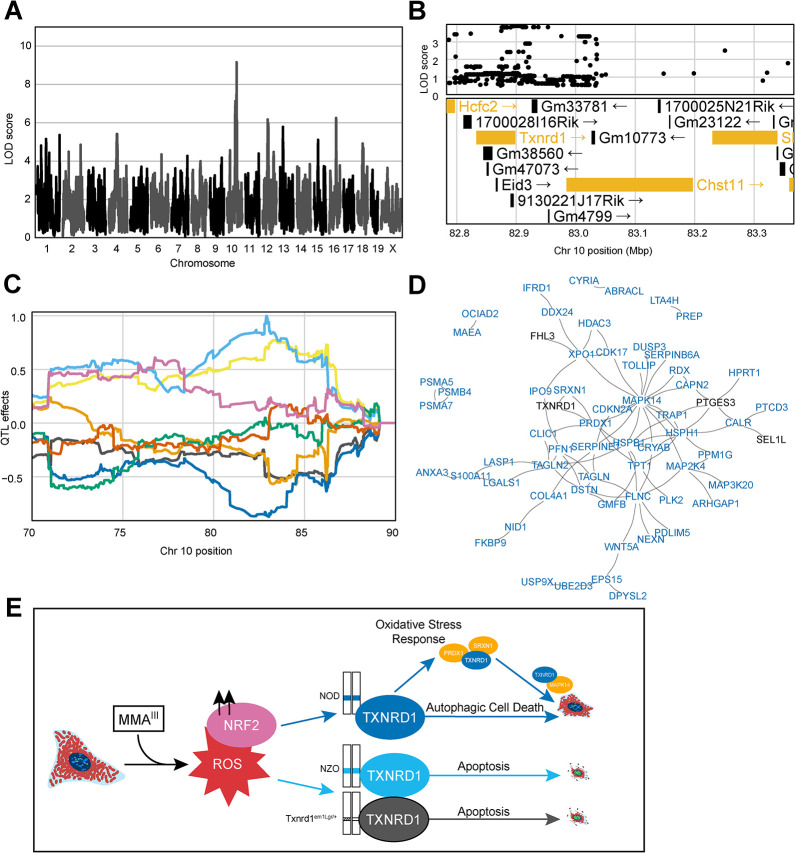
Fig 6. Noncoding Variation in Txnrd1 Modulates MMAIII-Induced Cell Death.
(A) QTL scan for the `γH2AX-negative cells slope Cell Area μm2 mean per well' cmQTL with the maximum peak at Chromosome 10: 82,906,780 bp (GRCm38) and a LOD score of 9.16. (B) Variant association mapping within the CI the cmQTL `γH2AX-negative cells slope Cell Area μm2 mean per well'. Top panel shows the LOD scores of the known, segregating variants in the 8 DO founders (GRCm38). Bottom panel shows the gene models within the respective CI. Each point represents a variant. Colors indicate whether a gene is expressed > 0.5 TPM (gold) or < 0.5 TPM (black). The arrow indicates the direction of transcription. (C) Haplotype effects plot showing the eight DO founders (colors, see Methods) for the `γH2AX-negative cells slope Cell Area μm2 mean per well' cmQTL across the surrounding region on chromosome 10 (Mbp). (D) String-db functional enrichment network of the significantly increased protein interactors detected using immunoprecipitation mass spectrometry (IP-MS) in DO fibroblasts with NOD alleles (n = 6) at the maximum locus for the `γH2AX-negative cells slope Cell Area μm2 mean per well' cmQTL exposed to 0 and 0.75 μM MMAIII concentrations. Colors indicate whether a protein, or node, was shared with a similar experiment in DO fibroblasts with the NZO allele (n = 5). Black represents shared TXNRD1 interactors, and blue represents unique NOD-TXNRD1 interactors. (E) Mechanistic summary of allele specific Txnrd1 responses across the NOD haplotype (blue), NZO haplotype (light blue), and heterozygous SECIS knockout model (Txnrd1em1Lgr/+). Our data suggest DO fibroblasts with the NOD allele have a more robust oxidative stress response upon MMAIII exposure, ultimately succumbing to autophagic cell death represented by increased cell size at medium MMAIII concentrations. In comparison, DO fibroblasts with the NZO allele the Txnrd1em1Lgr/+ alleles exhibit morphology consistent with apoptosis as shown by brighter Hoechst 33342 labeling and smaller cells.