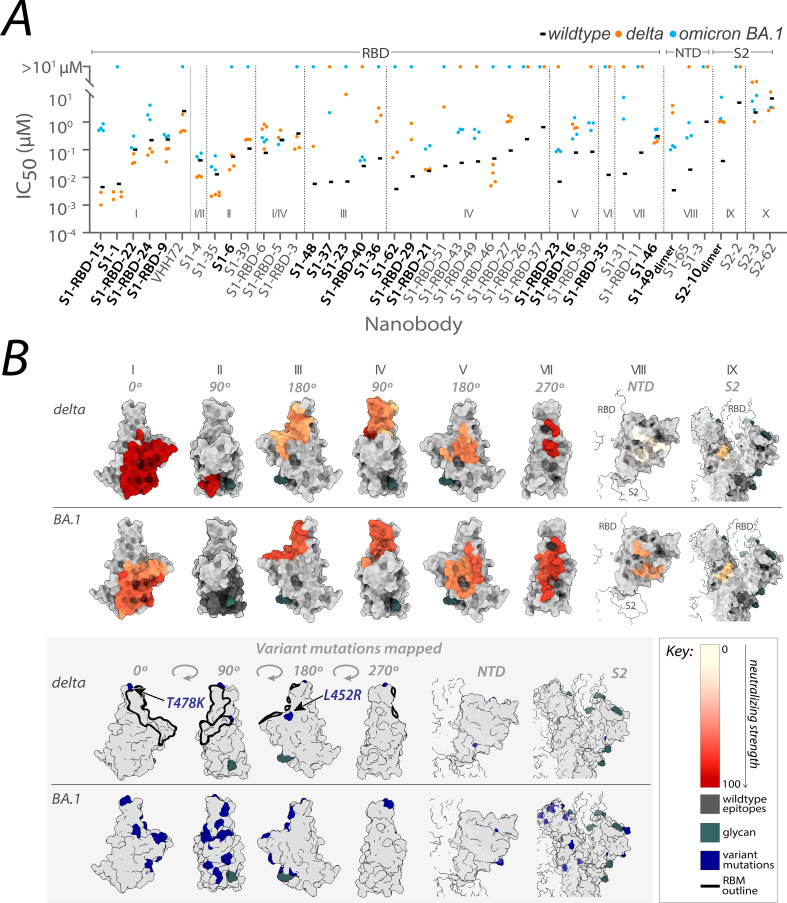
Figure 1. Nanobody repertoires generated against wild-type SARS-CoV-2 remain efficacious.
Nanobodies targeting the S1-RBD, S1 non-RBD, and S2 regions of spike effectively neutralize lentivirus pseudotyped with delta and omicron BA.1 SARS-CoV-2 spikes (PSV) from infecting angiotensin-converting enzyme 2 (ACE2)-expressing HEK293T cells. (A) The half-maximal inhibitory concentration (IC50) is reported for the indicated nanobodies against wild-type (Mast et al., 2021), delta, and omicron BA.1 PSV. These values are summarized in Figure 2—source data 1. Nanobodies are grouped by epitope and arranged within each epitope by neutralization efficacy against the wild-type PSV. n ≥ 4 (B) The structural differences in the receptor-binding domain (RBD) of the delta (PDB ID: 7SBO) and omicron BA.1 (PDB ID: 7T9K) variants are depicted. Nanobody epitopes are heat-mapped ranging from pale white (epitopes with weak neutralization against SARS-CoV-2) to dark red (indicating strong neutralization). Boxed in gray are mutations specific to each variant mapped in blue on the aforementioned structures. The nanobodies that contributed to epitope mapping are in bold in panel A. The color bar scale for each epitope is the neutralizing strength of each nanobody epitope, calculated as the normalized −log10 ratio of nanobody binding (IC50) to variant versus wild-type SARS-CoV-2 Spike S1. For groups with multiple nanobodies, the average −log10 (IC50) is first calculated for the nanobodies within that group, then normalized to a neutralization score within the 0–100 range using the min and max average −log10 (IC50) for that group. A higher score indicates more potent neutralization of the variant relative to the wild-type. All structural representations were created on ChimeraX (Pettersen et al., 2021).