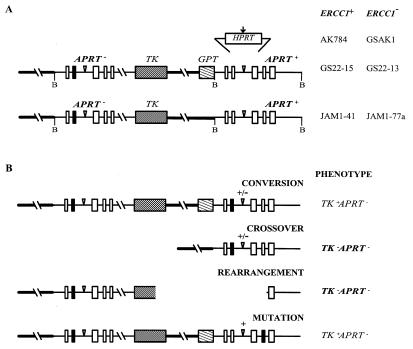
Figure 1.
Molecular structures of the substrates at the APRT locus in ERCC1+ and ERCC1– cell lines and of the products isolated in various selections. (A) The inserted HPRT sequence is shown above its site of insertion into the second intron of the downstream, functional APRT gene (the five exons of APRT are shown as boxes). The vertical arrow indicates the location of the I-SceI recognition site in the HPRT insert. Inverted triangles indicate the positions of the FRT recognition sequences. The upstream copy of APRT is non-functional by virtue of a truncated exon 5 and a single base substitution in exon 2 (filled box). The upstream and downstream copies share 6.8 kb of homology; 4.5 kb upstream of the APRT gene (thick line) and 2.3 kb of homology within the gene itself. Cleavage sites for the restriction enzyme BamHI (B), which was used in Southern analyses, are indicated. The hybridization probe corresponds to the downstream BamHI fragment that encompasses the APRT gene, but included no inserted sequences. (B) Types of possible APRT– products. Only TK–APRT– products (bold) were analyzed in these experiments; they were identified based on Southern patterns after BamHI cleavage and PCR analysis. Crossovers yielded a single BamHI cleavage fragment of 4.0 kb for JAM1-41 and JAM1-77a and a single BamHI cleavage fragment of 4.0 or 4.8 kb, depending on whether the insert was lost or retained, for AK784 and GSAK1 (+/– indicates presence or absence of the insert). Rearrangements gave a Southern pattern that did not correspond to crossovers (or any other class of products).