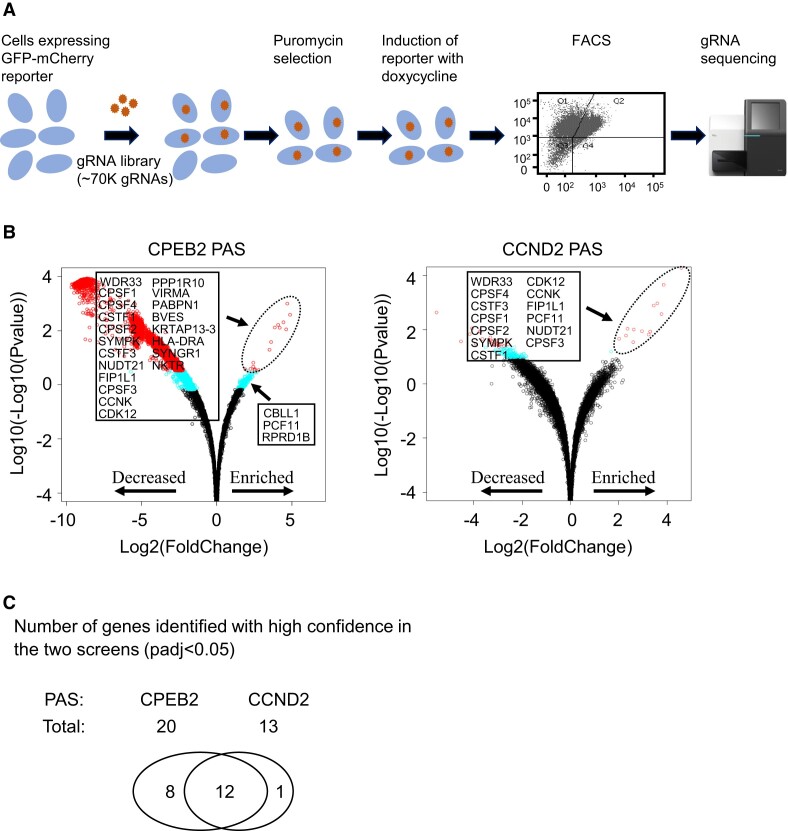
Figure 2.
CRISPR/Cas9 screens identified CPA factors. (A) CRISPR/Cas9 screening pipeline. Flp-In T-REx HEK293 cells expressing the GFP-mCherry readthrough reporter with an inserted PAS were infected with the pooled lentiviral human TKOv3 gRNA library followed by puromycin selection. After doxycycline-induced expression of the two reporters, cells were sorted by FACS. Genomic DNA was isolated from unsorted cells and sorted cells that exhibited an increased mCherry/GFP ratio, and the gRNAs were amplified and subjected to sequencing. (B) CRISPR/Cas9 screen results for HEK293 cell lines expressing the GFP and mCherry reporters separated by the distal PAS from the CPEB2 (left) and CCND2 (right) genes. Scatterplots (volcano plots) display pairwise comparisons of the gRNAs between sorted cells that exhibited an increased mCherry/GFP ratio and unsorted cells. Red circles: Padj < 0.05. Blue circles: Padj < 0.25, but >0.05. Black circles: Padj > 0.25. Experiments were performed in biological quadruplicate. Padj values were determined using the Benjamini–Hochberg adjustment for multiple comparisons. (C) Genes identified in both screens with high confidence. The gRNAs of 12 genes were enriched with high confidence (Padj < 0.05) in sorted cells with increased mCherry/GFP ratio as compared with unsorted cells in both screens.