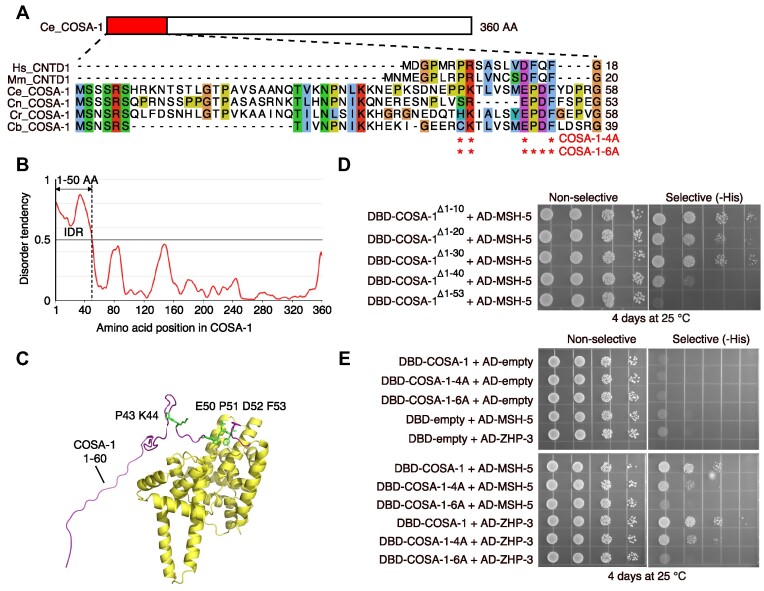
Figure 2.
The N-terminus of COSA-1 is required for interaction with MSH-5 and ZHP-3. (A) Schematic of the N-terminal sequences from COSA-1 and its orthologs from different organisms. The sites of mutations are indicated. Sequences are shown as follows: Hs, Homo sapiens (NP_755749.2, NCBI Sequence ID); Mm, Mus musculus (NP_080838.1, NCBI Sequence ID); Ce, Caenorhabditis elegans (NP_497607.3, NCBI Sequence ID); Cn, Caenorhabditis nigoni (PIC38620.1, GenBank); Cr, Caenorhabditis remanei (XP_053587050.1, NCBI Sequence ID); Cb, Caenorhabditis brenneri (EGT33696.1, GenBank). (B) The intrinsically disordered region (IDR) of COSA-1 was analyzed by IUPRED3 and the propensity for disorder was drawn as a solid line. Values above 0.5 indicate an IDR. (C) The structure of COSA-1 predicted by Alphafold2 revealed a long flexible N-terminus. The sites of mutations are indicated. (D) The yeast two-hybrid system was used to further refine the region of COSA-1 required for the interaction with MSH-5. (E) Analysis of interaction between COSA-1-4A or 6A and ZHP-3 or MSH-5 by yeast two-hybrid system.