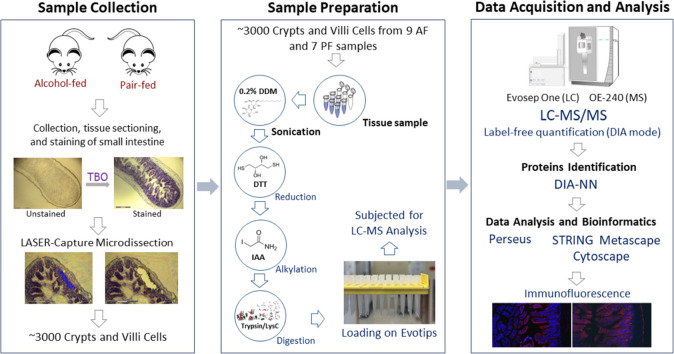
Figure 1.
Schematic illustration of the workflow for the LCM-based proteomics study. The cells of crypts and villi regions (equivalent to ∼3000 cells) were dissected from alcohol-fed and pair-fed mouse small intestine samples using LCM. Each scale bar within the image corresponds to 300 μm. The collected tissue samples were subsequently subjected to in-solution digestion and loading onto Evotips for further analysis by using LC–MS/MS. The output raw files underwent rigorous protein identification and comprehensive statistical and bioinformatic analyses to unravel the underlying molecular mechanisms. The expression patterns of some essential proteins were further validated with immunofluorescence imaging.