Figure 4.
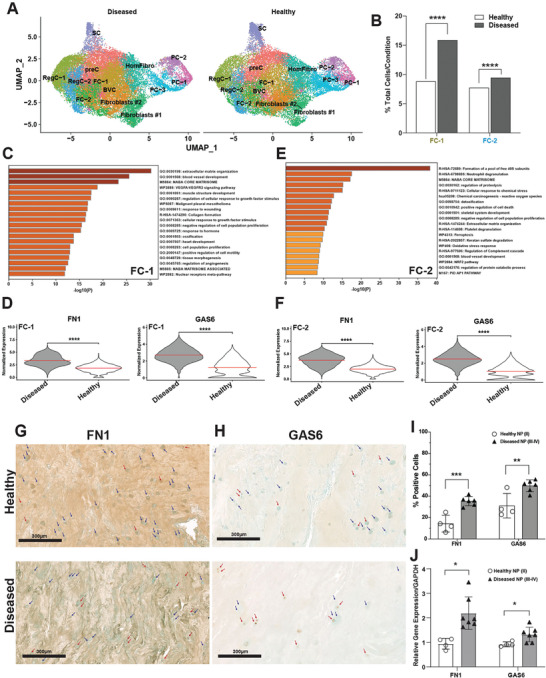
Expansion of fibrotic clusters in diseased NP. A) Visualization of clustering by split UMAP plot of healthy (n = 3) versus diseased (n = 8) NP samples. B) Quantification of FC‐1 and FC‐1 subsets in healthy versus diseased NP. Data are shown as percentage of total cells for each condition. ****p<0.0001 by comparison of proportions tests. (C,E) Metascape analysis of gene markers of FC‐1 C) and FC‐2 E). (D,F) FN1 and GAS6 are among the top DEGs in diseased NP compared to healthy in FC‐1 D) and FC‐2 F). G,H) IHC for FN1 (G) and GAS6 (H) was performed on healthy (grade II, n = 4) and diseased (grade III‐IV, n = 6) NP tissues. Counterstaining was performed using methyl green to calculate percentages of positive cells verses total cell numbers. Red arrows indicate examples for positive cells and blue arrows show negative cells. Scale bars indicate 200 µm. I) Quantifications of cells positive for FN1 and GAS6 in healthy (n = 4) versus diseased (n = 6) NP are shown. Two‐tailed unpaired Student's t‐test were used to establish statistical significance. Data are expressed as means ±SD (standard deviation).** p <0.01, *** p <0.001. J) qPCR showing FN1 and GAS6 mRNA abundance in whole healthy (grade II, n = 4) versus diseased (grade III‐IV, n = 7) NP. Data are relative to GAPDH. Two‐tailed unpaired Student's t‐test were used to establish statistical significance. Data are expressed as means ±SD (standard deviation). *p <0.05.
