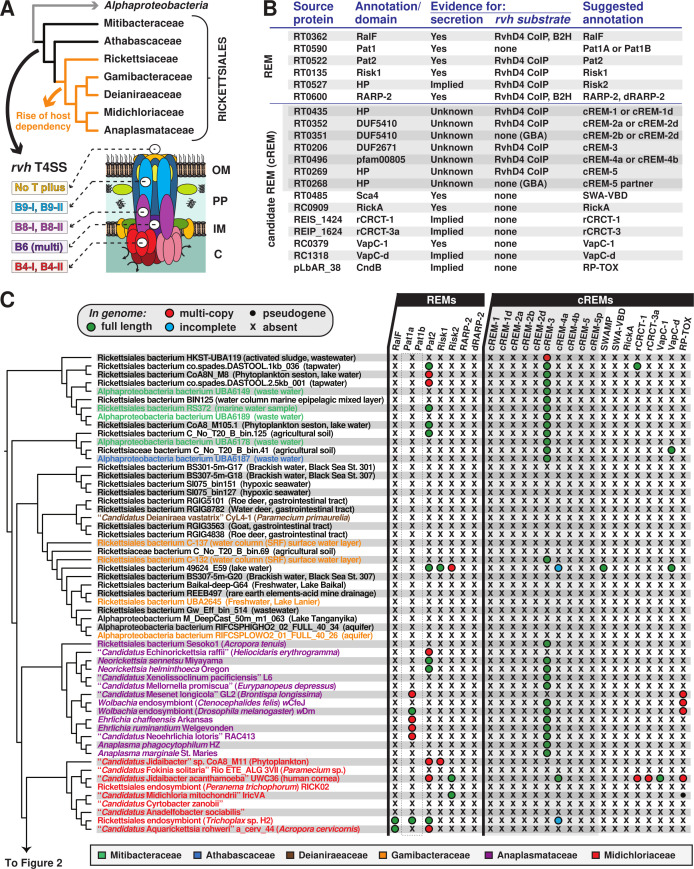
Fig 1.
Probing Rickettsiales diversity for the evolution of Rickettsia type IV secretion system effectors. (A) The atypical Rickettsiales vir homolog (rvh) T4SS is a hallmark of Rickettsiales that was present before the origin of host dependency (orange) (28, 34). Schema depicts recent genome-based phylogeny estimation (28). Rvh characteristics (38, 45) are described at the bottom and further in Fig. S1. (B) List of Rickettsia rvh effector molecules and candidate REMs. GBA, guilty by association (meaning a tandem gene with sequence similarities has experimental support for encoding a REM). “Implied” means analogous proteins are known to be secreted by other bacteria and/or the effector has strongly predicted host cell targets. Secretion, coimmunoprecipitation (CoIP), and bacterial two-hybrid (B2H) data are compiled from prior reports (56–58, 74–81). SWA, Schuenke-Walker antigen domain. (C) Phylogenomic analysis of Rickettsia REMs and cREMs in non-Rickettsiaceae lineages. The cladogram summarizes a phylogeny estimated from concatenated alignments for RvhB4-I and RvhB4-II proteins from 153 rickettsial assemblies (full tree, Fig. S2; sequence information, Table S1). Non-Rickettsiaceae lineages are shown (see Fig. 2 for Rickettsiaceae). The dashed box for Pat1 proteins indicates the inability to confidently discern Pat1a and Pat1b homology outside of Tisiphia and Rickettsia species (see Fig. 2 and 4). SWAMP, SWA modular proteins. Information for all REMs and cREMs is provided in Table S2.