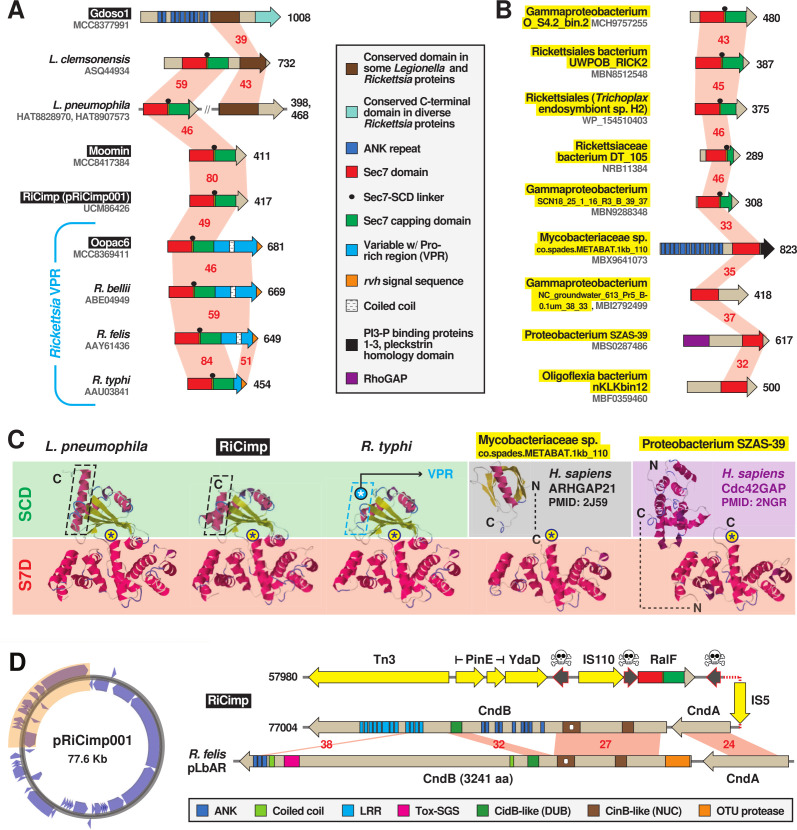
Fig 3.
MAG analysis divulges novel diversity of bacterial Sec7-domain-containing proteins. Black boxes provide short names for MAGs from Davison et al. (73). These and additional newly discovered RalF-like proteins (highlighted yellow) substantially expand the prior recognized RalF diversity (56, 84, 88, 90). Structural models for proteins are found in Fig. S4D through F. (A and B) Insight from (A) novel Legionella and rickettsial architectures and (B) diverse RalF-like proteins discovered in MAGs. Red shading and numbers indicate percent aa identity across pairwise alignments (sequence information in Table S2). All protein domains are described in the gray inset. (C) Comparison of the Legionella pneumophila RalF structure (PDB 4C7P) (88) with predicted structures of S7D-SCD regions of RiCimp RalF (LF885_07310) and Rickettsia typhi RalF (RT0362), and S7Ds of Mycobacteriaceae sp. co.spades.METABAT.1kb_110 (K2 × 97_15435) and Proteobacterium SZAS-39 (JSR17_09325). The delineation of the Sec7 domain (S7D, red) and Sec7-capping domain (SCD, green if present) is shown with an approximation of the active site Glu (asterisk). Additional eukaryotic-like domains for the non-rickettsial proteins are noted. Modeling was done with Phyre2 (95). More detailed structural explanation can be found in Fig. S4C. (D) RiCimp plasmid pRiCimp001 carries RalF and a CindB/A toxin-antidote module similar to those characterized or implicated in reproductive parasitism (94). Gene region drawn to scale using the PATRIC compare region viewer tool (96). Yellow, transposases and other mobile elements; skull-and-crossbones, pseudogenes; other domains are described in the gray inset at the bottom. Plasmid map created with Proksee (https://proksee.ca/).