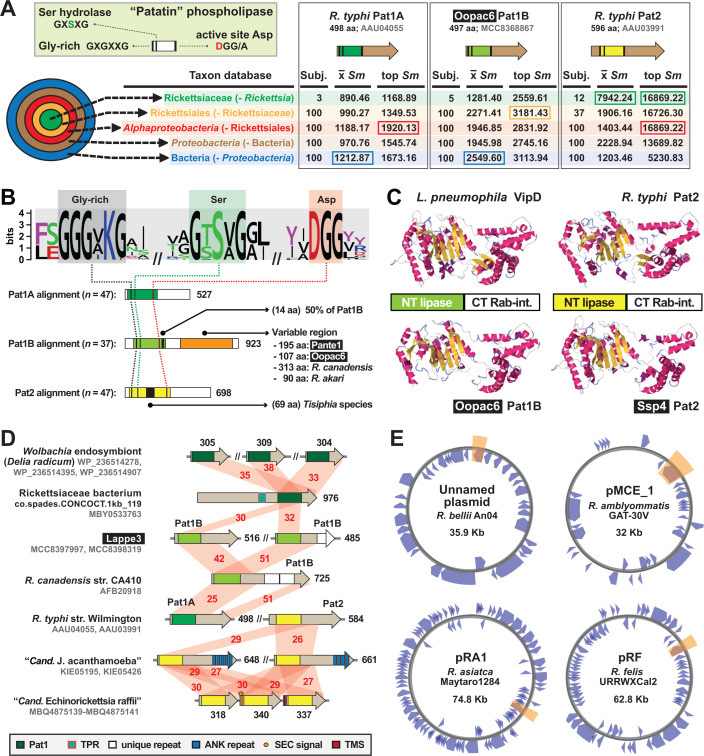
Fig 4.
Divergent patatin phospholipases are recurrent in rickettsial evolution. Black boxes provide short names for MAGs from Davison et al. (73). (A) PLA2 active site characteristics and divergent patatin forms. Green inset describes general patatin domain and active site architecture. HaloBlast results for Pat1A, Pat1B, and Pat2 (query sequences described at the top) are shown, with top-scoring halos boxed (full results in Table S4). (B) Sequence logo (103) showing conservation of the PLA2 active site motifs across Tisiphia and Rickettsia patatins (sequence information provided in Table S2). Pat1A, Pat1B, and Pat2 sequences were aligned separately with MUSCLE (104) (default parameters) with active site motifs compiled for conservation assessment. Features unique to each patatin are noted. (C) Legionella pneumophila VipD structure (PDBID: 4AKF) and modeling of three rickettsial patatins to VipD using Phyre2 (95). (D) Diverse architectures for select patatins. Red shading and numbers indicate percent aa identity across pairwise alignments (sequence information in Table S2). All protein domains are described in the gray inset. Dark green indicates Pat1 domains not grouped into A or B. (E) Four Rickettsia plasmids carry pat1B (shaded orange). Plasmid maps were created with Proksee (https://proksee.ca/).