Figure 1 |. The uLIPSTIC system.
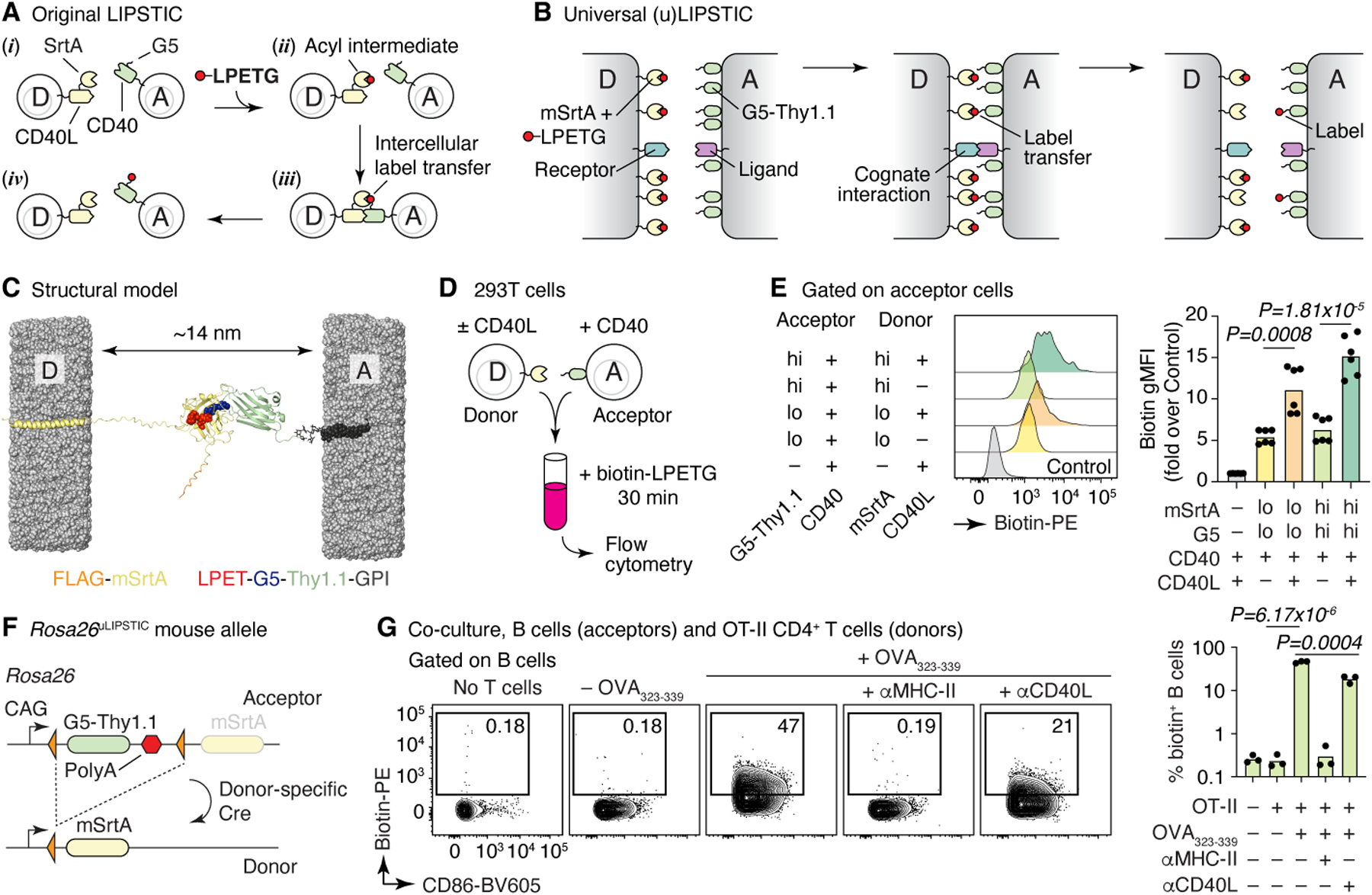
(A, B) Schematic comparison of the original2 and universal LIPSTIC systems. In the original system (A), SrtA and G5 were brought into proximity by fusion to a receptor–ligand pair involved in a cell–cell interaction, allowing intercellular transfer of labeled substrate (LPETG) from donor cell “D” to acceptor cell “A.” In uLIPSTIC (B), SrtA and G5 (fused to the irrelevant protein Thy1.1) are anchored non-specifically to the cell membrane at high density; the enzymatic reaction is allowed to proceed when apposing membranes come within a short distance (< 14 nm) of each other, which can be driven by interactions between any receptor–ligand pair of the appropriate dimensions. (C) Computational model depicting the inter-membrane span of fully extended mSrtA upon transfer of the LPETG substrate onto G5-Thy1.1. (D,E) Populations of 293T cells co-transfected with high or low levels of either mSrtA or G5-Thy1.1 were co-incubated in the presence of biotin-LPETG for 30 min and analyzed by flow cytometry. Histograms show the extent of labeling of acceptor cells. Each symbol on column plot represents one technical replicate, pooled from two independent experiments. (F) Rosa26uLIPSTIC allele. Using the Ai9 high-expression backbone20, a LoxP-flanked G5-Thy1.1 is followed by mSrtA. Cre-recombinase switches cells from “acceptor” (G5-Thy1.1+) to “donor” (mSrtA+) modes. (G) Rosa26uLIPSTIC/+.CD4-Cre OT-II donor T cells were co-cultured with Rosa26uLIPSTIC/+ acceptor B cells in the presence or absence of OVA323–339 peptide and blocking antibodies to CD40L and MHC-II. Flow cytometry plots show biotin-LPETG transfer from T to B cells. Each symbol in column plot represents a biological replicate from three independent experiments. For (E) and (G), P-values were calculated using two-tailed Student’s tests.
