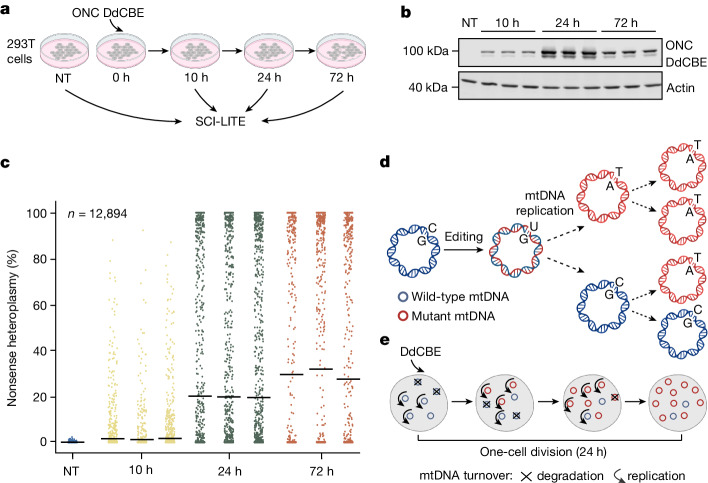
Fig. 2. mtDNA base editing leads to a bimodal distribution of heteroplasmy.
a, Schematic overview of the SCI-LITE experiment. 293T cells were transfected with the ONC DdCBE, introducing a nonsense mutation in the MT-ND4 gene. Cells were cultured for 10 h, 24 h and 72 h, harvested and used for SCI-LITE. b, Western blot of ONC DdCBEs, showing expression of FLAG-tagged DdCBE halves (see Supplementary Fig. 2 for uncropped images). Actin was used as a loading control. n = 3 biological replicates are shown. c, Single-cell heteroplasmy in 293T cells interrogated using SCI-LITE. n = 3 biological replicates of edited cells are shown. Lines represent the mean for single biological replicates. Dots represent single cells. NT, not treated. d, Proposed model for heteroplasmy installation using mtDNA base editing. DdCBEs convert cytosine to uracil within double-stranded mtDNA molecules. Replication of one edited mtDNA molecule leads to the formation of one mutated and one wild-type mtDNA molecule. Further editing and replication of wild-type mtDNA molecules leads to formation of 50% mutant and 50% wild-type molecules. Replication of mutated molecules results in 100% mutant mtDNA molecules. e, For a single cell, assuming a fully active editor, after n rounds of mtDNA replication, we would expect that the heteroplasmy of the cell H = 1 − (1/2)n. Under this simple model, after just a few rounds of mtDNA replication in the face of an active base editor, a single cell will achieve a heteroplasmy level approaching 100%, but only if the mtDNA replication and turnover rate exceeds the cell division rate. Schematics in parts a,d,e were created using BioRender (https://biorender.com).