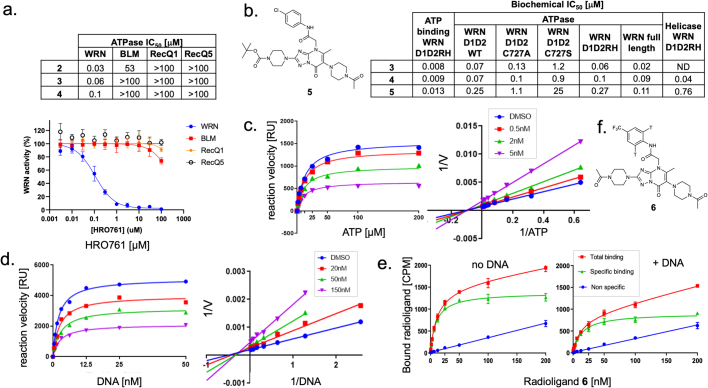
Extended Data Fig. 2. HRO761 and related WRN inhibitors are selective WRN inhibitors mixed competitive with ATP and uncompetitive with DNA.
a. Biochemical ATPase IC50 activity and concentration response curves of the WRN inhibitors HRO761 (4), and analogues 2 and 3 on WRN as well as RecQ helicases BLM, RecQ1 and RecQ5. Data represent mean of quadruplicates expressed as %inhibition ± SD. b. Comparison of biochemical ATP binding, ATPase and helicase IC50 across different WRN constructs and mutants (reported IC50 values in a and b are the geometrical means of at least 2 independent experiments., structure of 5 shown on the left.). c. Michaelis-Menten plots of the WRN ATPase assay (0.5 nM WRN D1D2RH, 3 nM ssDNA FLAP26, 50 mM NaCl) at varying concentrations of 4 (for legend, see inset) revealed a non-competitive or mixed mode of action (Lineweaver-Burk plot on the right), interpreted as mixed ATP-competition due to competition in ATP binding assay (b.) d. KDNA determination in the ATPase assay (0.5 nM WRN D1D2RH, 100 µM ATP, 150 mM NaCl, ssDNA: FLAP26) at varying concentrations of 5 (for legend, see inset) revealed a non-competitive or mixed mode of action (double reciprocal plot on the right), interpreted as a non-competitive mode of action due lack of DNA competition in a radioactive binding assay (e.). For c, d, Data represent initial Kobs determined by following the ATPase reaction for 20 min. e. Saturation binding experiments of radiolabeled 6 (structure shown in f.) performed with 10 nM WRN (D1D2RH) in absence (left) or in presence of 50 nM dsDNA (right) using a Scintillation Proximity Assay resulted in a similar Kd (10/14 nM). Data points represent the mean of triplicate ±SD (each experiment was repeated at least twice).