Fig. 1.
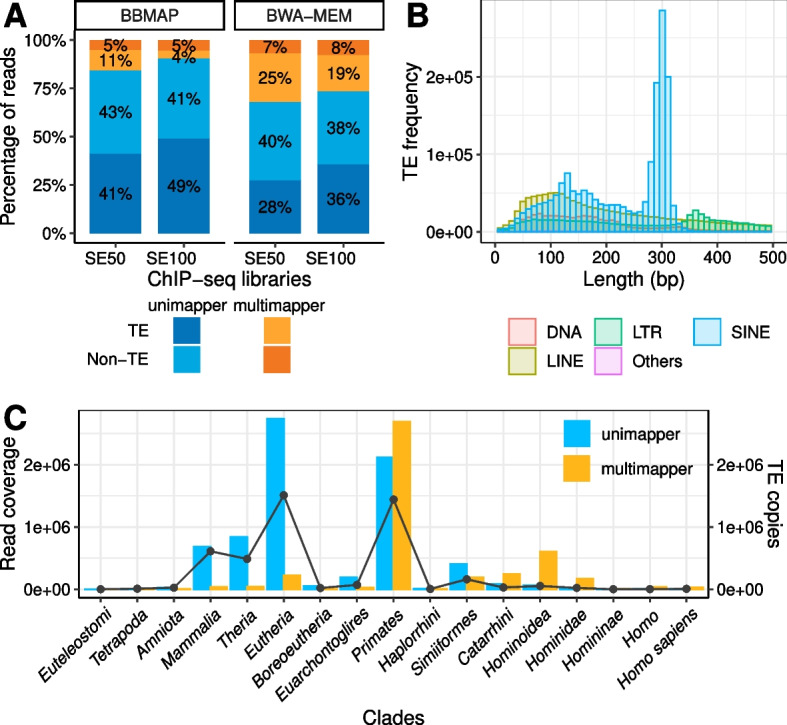
Discarding multimappers leads to epigenetic mischaracterization of young TEs. A Percentage of uni- and multimapper reads mapping to portions of the human genome annotated as TEs and not annotated as TEs (non-TE) for dataset 1 containing two ChIP-seq libraries generated by the ENCODE consortium using single-end 50 bp (“SE50”) and 100 bp (“SE100”) reads. Mapping was performed with two different mapping tools: with BBMap and Bwa mem. TEs are the major source of ChIP-seq multimappers in the human genome. B Length distribution of TE individual copies. Only TEs shorter than 500 bp are shown. Note that ~ 74% (856 out of 1,160) of the TEs in the human genome are longer than 500 bp, spanning up to 153,104 bp. The bin width is 10 bp. TEs were classified as DNA, LTR (long terminal repeat), SINE (short interspersed nuclear element), LINE (long interspersed nuclear element) and Others (e.g., rolling-circle (RC), unknown classification). Standard NGS reads are too short to fully cover most TE copies, explaining why TEs often give rise to multimappers. C Read coverage (bar plot on the left y-axis; see Methods) per clade for the SE100 ChIP-seq library (dataset 1) for uni- and multimappers. Reads were mapped using Bwa mem. TEs found in multiple clades (e.g., L1HS and L1P1) were assigned to the “younger” clade (Homo and Hominoidea, respectively). Only 30 out of 1,160 different TEs in the human genome (~ 3%) have not been annotated to any clade and were not represented. The number of TE copies for each clade (line plot on the right y-axis) shows that the majority of TEs are Primates and Eutherian-specific. Evolutionary young TEs are prone to be underrepresented when excluding multimappers from ChIP-seq analysis
