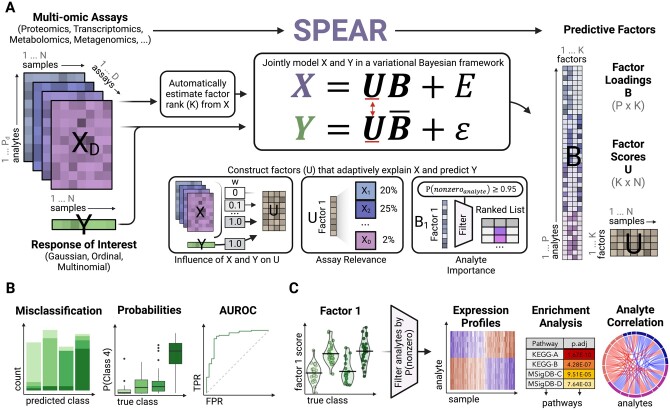
Figure 1.
SPEAR workflow overview: (A) SPEAR takes multi-omics data (X) taken from the same N samples, as well as a response of interest (Y). SPEAR supports Gaussian, ordinal, and multinomial types of responses. From these inputs, the algorithm first automatically estimates the minimum number of factors to use in the SPEAR model. X and Y are then jointly modeled in a variational Bayesian framework to adaptively construct factor loadings (B) and scores (U) that explain variance of X and are predictive of Y (reflected in ). (B) SPEAR factors are used to predict Y and provide probabilities of class assignment for ordinal and multinomial responses. (C) Downstream biological interpretation of factors is facilitated via automatic feature selection, expression profile analysis, enrichment analysis, and analyte correlation