Figure 4.
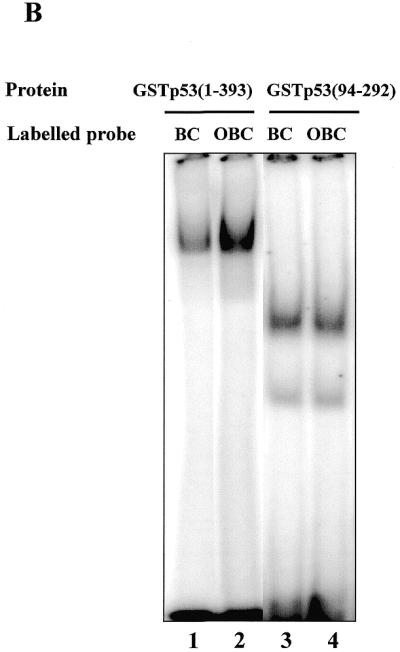
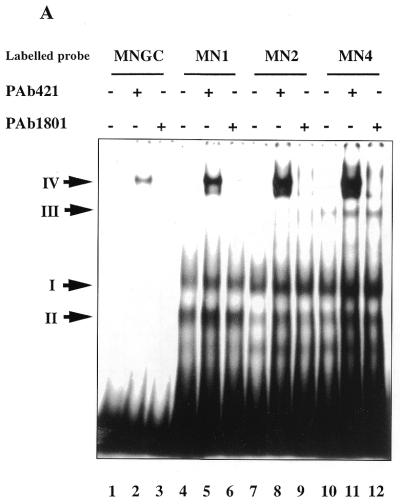
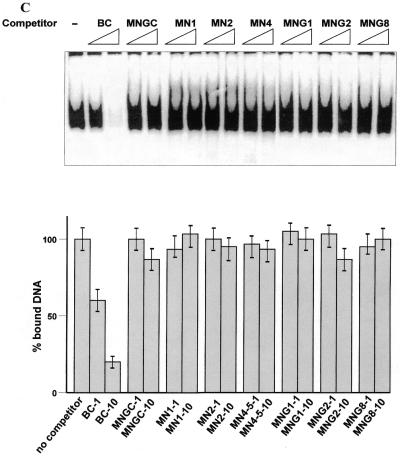
(A) Interaction of the C-terminus with protruding DNA ends induces the core domain–DNA interaction. Labelled dsDNA oligonucleotides MNGC (blunt end) and MN1, MN2 and MN4, with 1, 2 and 4 nt single-stranded ends, respectively, were used. C-terminal-mediated complexes I and II and PAb421-induced core domain-mediated complex IV are indicated by arrows. Efficiency of the p53 core domain–DNA interaction with dsDNA increases with the length of protruding ends on the DNA probe. (B) The presence of protruding single-stranded ends enhances specific DNA binding by the core domain in cis. Interaction of full-length p53 with labelled blunt end probe BC, containing the p53 binding site (lane 1), was less efficient than interaction with the staggered end OBC probe which has the same sequence (lane 2). The isolated p53 core domain protein (residues 94–292) bound both oligonucleotides with the same efficiency (lanes 3–4). In order to visualise the p53–BC complex in the absence of PAb421 antibody, 50 ng of p53 protein was used. (C) Isolated core domain protein p53(98–292) does not recognise protruding ends or gaps on dsDNA. (Upper) Specific oligonucleotide BC was used as a labelled probe and cold double-stranded oligonucleotides with different structures were used as competitors. (Lower) PhosphorImager-assisted quantitation of the series of three competition experiments. No statistically significant difference in efficiency of challenging core–BC complexing was observed between double-stranded oligonucleotides with internal gaps (MNG1, MNG2 or MNG8), staggered ends (MN1, MN2 or MN4) or a blunt end (MNGC).