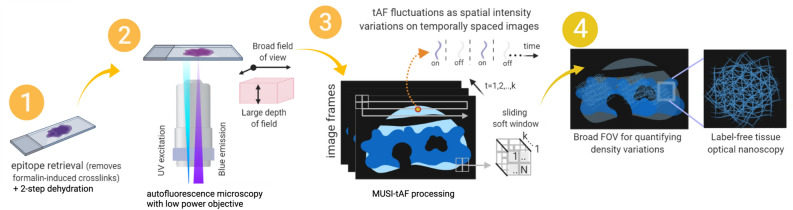
Figure 1.
Schematic diagram for MUSI-tAF workflow. The FFPE tissue sections replete with autofluorescing matrix proteins are mounted on glass slides, deparaffinized and dehydrated. Epitope retrieval is performed to remove unwanted formalin-linked fluorescence. A low magnification epifluorescence microscope (20 , 0.80 NA objective) that allows a broad field of view (FOV) of m2 and a thicker depth of field (DOF) of m is used to acquire image stacks at excitation and emission wavelengths specific to matrix protein autofluorescence. The rapidly acquired image stacks are processed by the multiple signal classification algorithm to achieve super-resolution as previously described29. The MUSI-tAF image presents quantifiable density variations on a broad field for a better clinical context along with the nanoscopy of matrix collagen protein.