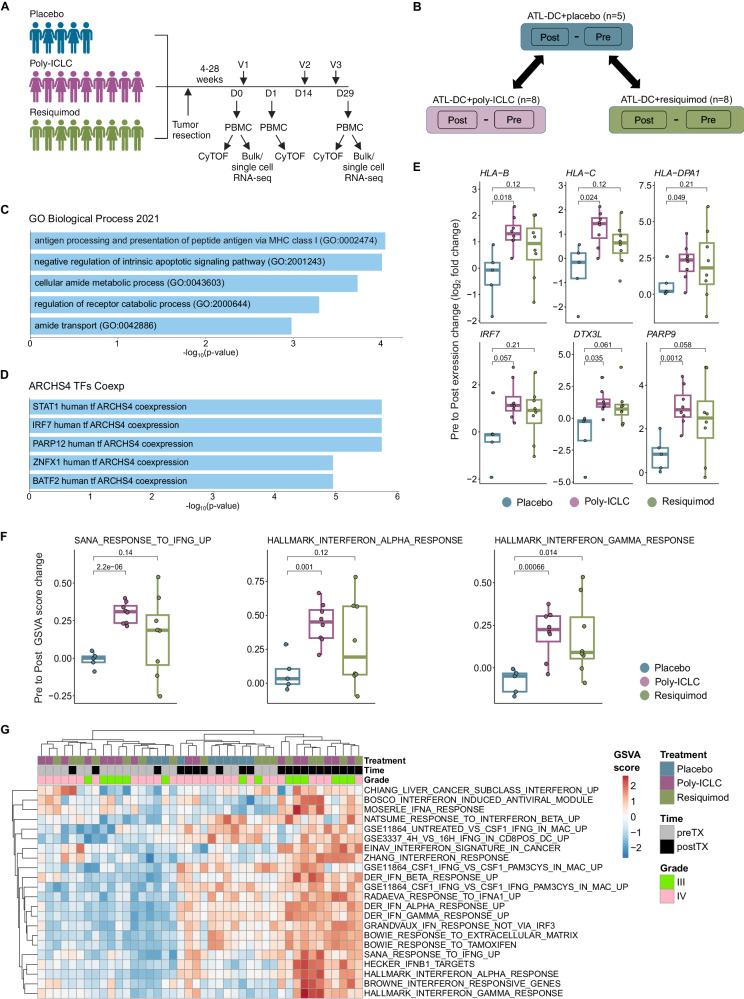
Fig. 1. Combination of ATL-DC vaccine and TLR agonists results in a robust interferon pathway activation in the patient PBMCs.
A Timeline of PBMC acquisition and analysis using CyTOF and/or RNAseq. V = vaccine, D = Day. (Figure created with the help of BioRender). B Schematic of differential gene expression analysis performed on pre-treatment and post-treatment PBMCs of indicated treatment groups. Differentially expressed genes (DEGs) in TLR agonist-treated groups are compared against their changes in the placebo group to identify DEGs specific to the TLR-agonist groups. C, D Enriched gene set terms in Gene Ontology Biological Process (C) or ARCHS4 TF Coexp (D) datasets that significantly overlap with the union of DEGs from ATL-DC + poly-ICLC and ATL-DC + resiquimod groups (P values, FDR-adjusted, two-sided fisher exact test). E Differential gene expression (pre vs. post-treatment fold change, in log2) of representative antigen presentation and IFN-related genes across treatment groups (P values, two-sided Welch t test). F Gene set enrichment score differences (pre vs. post-treatment, delta GSVA score) of representative IFN-related genesets across treatment groups (P values, two-sided Welch t test). G Heatmap of single-sample, gene set enrichment scores (GSVA) of type I and type II interferon genesets in pre-treatment, ATL-DC + placebo, ATL-DC+poly-ICLC and ATL-DC+resiquimod samples. The number of sample pairs analyzed in panels E and F are: ATL-DC+placebo, 5 pairs; ATL-DC+poly-ICLC, 8 pairs; ATL-DC+resiquimod, 8 pairs. The rectangular box in each boxplot represents the interquartile range (IQR), spanning from the first quartile (25th percentile, bottom of box) to the third quartile (75th percentile top of box). Inside the box, the median (50th percentile) is marked. The whiskers (shown as lines extending from the box) extend to the largest and smallest non-outlier values within 1.5 times the IQR, while outliers lie beyond the whiskers.