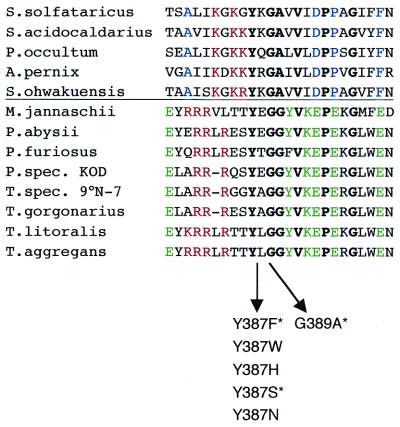
Figure 1.
Multiple sequence alignment of a 24–25 amino acid region surrounding the Y-GG/A motif in archaeal B-type DNA polymerases. In addition to the conserved motif Y-GG/A itself, V, P and G residues (in bold) are also conserved throughout the archaeal enzymes. A clear distinction can be made between crenarchaeal and euryarchaeal sequences (above and beneath the horizontal line, respectively). Residues conserved in euryarchaeal B-type DNA polymerases are marked in green, residues characteristic for crenarchaeal B1-DNA polymerase sequences are shown in blue. Another interesting feature is a cluster of positively charged amino acids (red) preceding the Y-GG/A motif in both archaeal subdomains, being Arg (R) in the case of the euryarchaeal sequences and Lys (K) in the case of the crenarchaeal ones. This cluster of lysines can also be found in eukaryal α-DNA polymerases in this region of the enzyme. Note also the high number of Glu (E) residues in the euryarchaeal sequences. The created Tag pol mutants are indicated under the alignment. Asterisks mark analogous mutants investigated in φ29 and Sso pol (2,3). Accession numbers of the Thermococcus spec. DNA polymerases: T.aggregans, O33845; T.litoralis, M74198; T.spec. 9°N-7, Q56366; T.gorgonarius, P56689. Accession numbers of the Pyrococcus spec. proteins: P.spec KOD, S71551; P.abyssii, P77916; P.furiosus, P80061; P.horikoshii, O59610; Methanococcus jannaschii, Q58295. Crenarchaeal species and accession numbers of the DNA polymerases: Sulfolobus solfataricus, P26811; Sulfolobus acidocaldarius, P95690; Pyrodictium occultum, D38573; Aeropyrum pernix, AB017500; Sulfurisphaera ohwakuensis, O50607.