Figure 3.
The interpretation pipeline, molecular diagnostic rates, and selected diagnostic findings from the iNeuron RNA-seq workflow
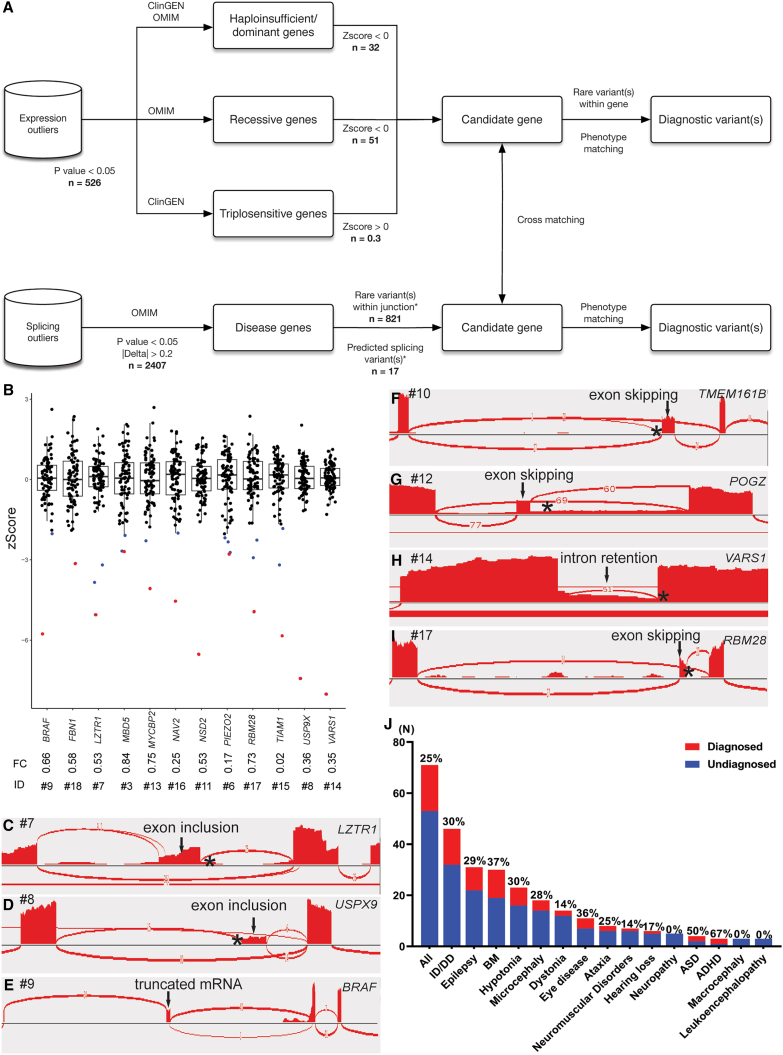
(A) Analytical and interpretation pipelines used in iNeuron RNA-seq analysis. The n values indicate the average number of variants passing filtration at each step.
(B) OUTRIDER identified 12 aberrant expression outliers as diagnostic findings (BRAF, FBN1, LZTR1, MBD5, MYCBP2, NAV2, NSD2, PIEZO2, RBM28, TIAM1, USP9X, and VARS1). Each dot represents an individual participant. The blue dots represent expression outliers that fulfill the analytical filtration standards defined in (A). The red dots represent molecular diagnostic findings with integrated considerations from analytical filtrations (blue dots), phenotypic matching, and correlation with DNA variant findings. For each molecular diagnostic expression outlier (red dot), its ID and expression fold change compared to controls are listed underneath.
(C–I) FRASER detected 10 aberrant splicing events, with 7 representative events (LZTR1, USP9X, BRAF, TEME161B, POGZ, VARS1, and RBM28) illustrated here. Aberrant junctions are shown in the Sashimi plot. The asterisk represents the position of the causal variant.
(J) Molecular diagnostic yields in individuals with different neurological phenotypes.