Figure 4.
Molecular diagnoses achieved by iNeuron RNA-seq through elevated expression of OMIM-N genes
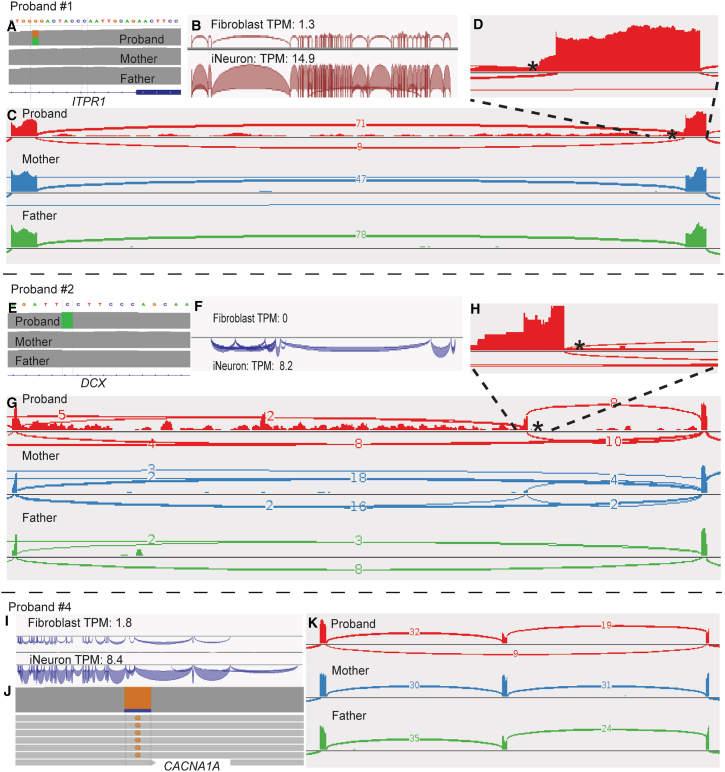
(A) Trio WGS reveals a de novo heterozygous intronic variant in ITPR1 (GenBank: NM_001378452.1; c.5980−17G>A).
(B) Significant increase in expression of ITPR1 after transdifferentiation.
(C) iNeuron RNA-seq demonstrates a 15-nucleotide retention from intron 45 in approximately 50% of the reads, as well as complete intron 45 retention in a small fraction of reads.
(D) Zoomed in view of the 15-nucleotide retention.
(E) Trio WGS identifies a novel hemizygous deep intronic variant (GenBank: NM_001195553.2; c.946+4588G>T) in DCX.
(F) DCX is not expressed in fibroblasts but is activated in iNeurons.
(G) Sashimi plot displays inclusion of a 13,549 bp cryptic exon from intron, positioned −31 bp from the intronic variant of DCX.
(H) Zoomed-in view of the new splice junction at the intron retention.
(I) Significant increase in expression of CACNA1A after transdifferentiation.
(J) Skewed expression of the variant allele in CACNA1A.
(K) Sashimi plot reveals exon skipping located 124 kb away from the missense variant. Variants are indicated by asterisks.