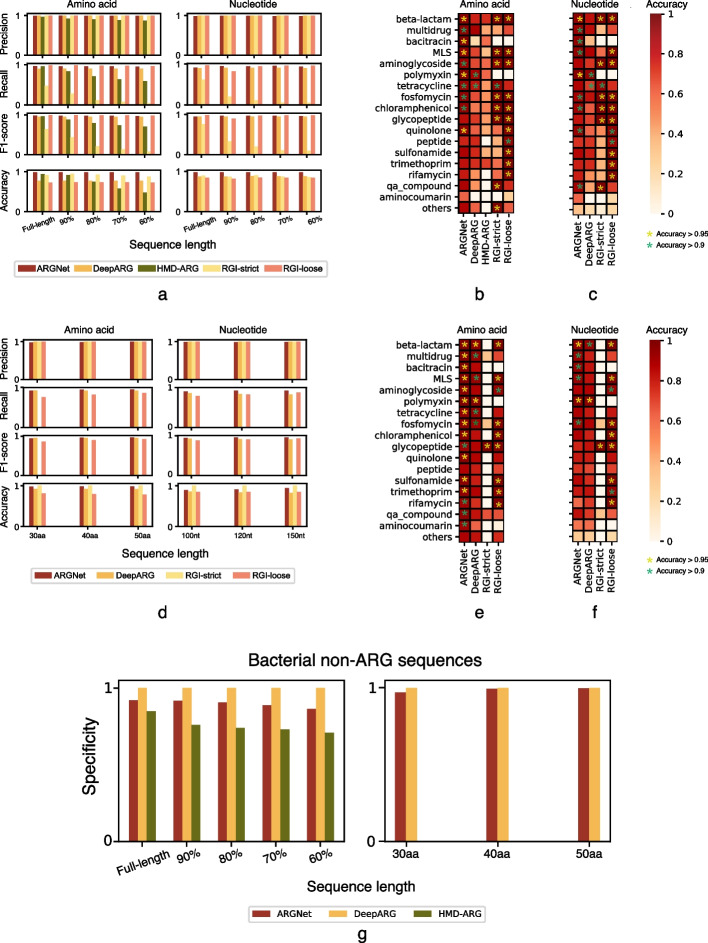
Fig. 5.
Performance of the deep learning models on test datasets. Precision, recall, F1 score, and classification accuracy of long sequences a and short sequences d. Left, amino acids; right, nucleotides. Datasets are indicated as proportion of the full-length sequences a and sequence length d. Classification accuracy of long amino acid sequences b, long nucleotide sequences c, short amino acid sequences e, and short nucleotide sequences f by antibiotic category. Nineteen minor ARG categories (with fewer than 50 sequences) are combined and displayed as one category called “others.” Accuracy level is indicated by asterisk with different colors. Identification specificity of bacterial non-ARG amino acid sequences g. Left, long sequence; right, short sequence. Datasets are indicated as proportion and sequence length, respectively