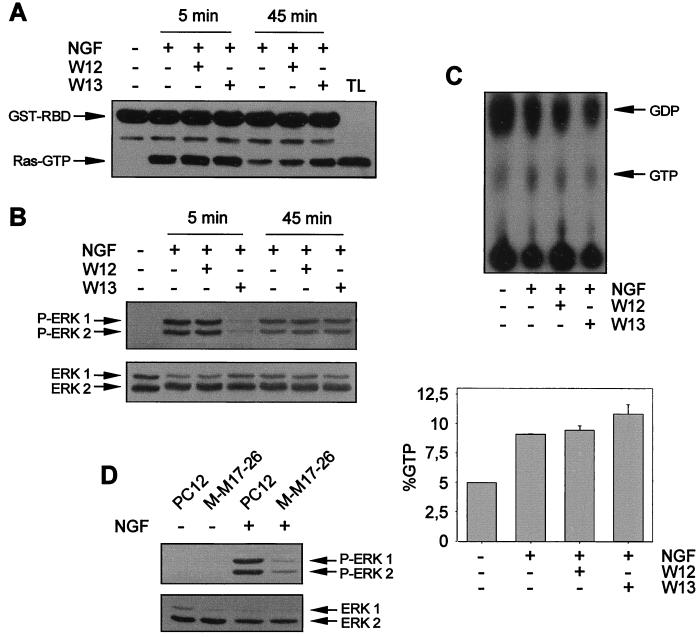
FIG. 5.
CaM inhibitors do not modify the profile of p21ras activation after NGF stimulation. PC12 cells were pretreated (+) or not pretreated (−) for 1 h with W12 or W13 and then stimulated (+) or not stimulated (−) for 5 or 45 min with NGF. After treatment, cells were lysed and protein extracts were obtained. (A) Protein extracts were subjected to precipitation with 50 μg of recombinant GST-RBD precoupled to gluthatione-Sepharose (see Materials and Methods). Precipitates were analyzed by Western blotting with an anti-pan-Ras antibody. Arrows indicate the positions of the proteins. TL, total cell extracts from PC12 cells. (B) Protein extracts from cell lysates in panel A were analyzed for ERK phosphorylation as described in the legend to Fig. 1. (C) PC12 cells were metabolically labeled with [32P]H3PO4, pretreated (+) or not pretreated (−) for 1 h with W12 or W13, and then stimulated (+) or not stimulated (−) for 5 min with NGF. Protein extracts were subjected to a Ras GTP-GDP loading assay as described in Materials and Methods (upper panel). The lower panel (graph) shows the average Ras activity from three independent experiments expressed as a percentage of GTP normalized by phosphorus according to the expression (GTP counts/3)/[(GTP counts/3) + (GDP counts/2)] × 100. (D) PC12 and M-M17-26 cells, which constitutively express the dominant negative Ha-ras mutant (Asn-17), were stimulated (+) or not stimulated (−) for 5 min with NGF. After treatment, cells were lysed and ERK phosphorylation was analyzed as described in the legend to Fig. 1.