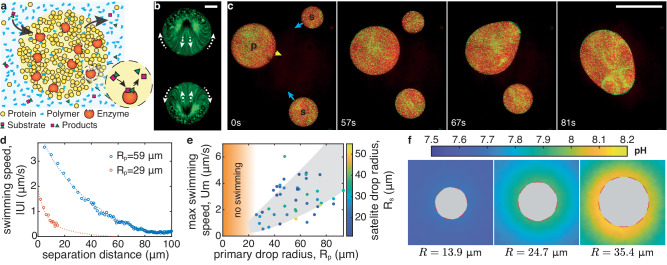
Fig. 1. Chemically active protein condensates swim toward each other.
a Schematic of a chemically active protein condensate. Polymers act as depletants, triggering condensation. Enzyme-rich droplets act as micro-chemical reactors. b Time projection over 1 min of fluorescent particles inside two adjacent chemically-active protein droplets catalyzing the urea-urease reaction (overall enzyme concentration ce = 0.6 μM, substrate concentration cs = 100 mM). The droplets are pinned on the surface (PEGDA 700). Arrows indicate the internal flow direction. Scale bar, 10 μm. c Image sequence of chemically active droplets on a non-wetting surface (PEGDA 12k gel, ce = 1.2 μM, cs = 100 mM, see Movie S1). The primary (p), satellite drops (s) and direction of motion are indicated in the first panel. Scale bar, 100 μm. d Swimming speed of two satellite drops, ∣U∣, as a function of the distance from their primary drops of radii Rp. Dashed curves are guides to the eye. e Maximum swimming speed, Um, as a function the primary drop radius, Rp, for 48 swimming-induced coalescence events (ce = 1.2 μM, cs = 100 mM). The color codes the satellite drop radius, Rs, the gray area is a guide to the eye. No swimming was observed for drops below 25 μm in radius as indicated by the orange area. f pH imaging around pinned reacting droplets of increasing size (ce = 1.2 μM, cs = 100 mM, PEGDA 700). The fluorescence signal inside the drops is masked by a gray disc, since they do not contain pH dye. Source data for (d, e) are provided as a Source Data file.