Abstract
Neutrophils are increasingly recognized as key players in the tumour immune response, and are associated with poor clinical outcomes. Despite recent advances characterizing the diversity of neutrophil states in cancer, common trajectories and mechanisms governing the ontogeny and relationship between these neutrophil states remain undefined. Here, we demonstrate that immature and mature neutrophils that enter tumours undergo irreversible epigenetic, transcriptional and proteomic modifications to converge into a distinct, terminally differentiated dcTRAIL-R1+ state. dcTRAIL-R1+ reprogrammed neutrophils predominantly localise to a glycolytic and hypoxic niche at the tumour core, and exert pro-angiogenic function that favors tumoral growth. We find similar trajectories in neutrophils present across tumour types and in humans, suggesting that targeting this deterministic program may provide an effective way to enhance cancer immunotherapy.
One-Sentence Summary:
Deterministic reprogramming of neutrophils drives their convergent differentiation in the tumoral niche
Neutrophils play significant roles in the immune response to infection and injury. As one of the first cells to enter a damaged site from the circulation, the rapid recruitment of large numbers of neutrophils into the tissue is key for their protective function (1). Notably, this process is co-opted in pathological settings such as cancer, where persistent neutrophil infiltration into the tumour has been consistently associated with poorer patient outcomes (2). Many studies have associated pro-tumoral functions to neutrophils, which in this context, have been referred to as granulocytic myeloid-derived suppressor cells (3). This uniform view has since been disrupted by the identification of a wide spectrum of neutrophil states in cancer, with differences in density (4, 5), surface marker (6–10), and transcript expression (11–13).
Neutrophil heterogeneity also exists in the bone marrow in the form of various maturation stages. Numerous studies have shown that granulocytic progenitor cells differentiate sequentially into precursor, immature and finally mature neutrophils, and each subset of cells possesses unique functional capabilities. (14–19). Notably, tumour-induced chronic inflammation triggers the premature egress of these precursors into the circulation and subsequently into the tumour (4, 14, 17, 19, 20), and extramedullary sites such as the spleen, (13, 14) have been proposed to be priming sites of pro-tumoral neutrophils (12, 21, 22). Collectively, this spectrum of neutrophil states, encompassing phenotypic and maturation differences, is proposed to make up the functional diversity of neutrophils in cancer (23–26).
Here, we utilize multi-omics approaches in pancreatic tumours at single-cell transcriptional and spatial resolution to examine neutrophil functional diversity in the context of their ontogenetic order, origin and influence from tissue signals. We find, unexpectedly, that although various populations of neutrophils enter the tumour, it is only inside the tumour that neutrophils enter a convergent trajectory that directs them towards a unique pro-tumoral state. Our findings challenge current models and reveal the potential of targeting neutrophils for cancer immunotherapy.
Intratumoral neutrophils converge into a distinct transcriptional state
To dissect neutrophil heterogeneity in cancer, we used an orthotopic model of pancreatic ductal adenocarcinoma (PDAC). Using a pancreatic cancer cell line previously established from a tumor in the Pdx1Cre; KrasG12D/+; Trp53R172H/+ (KPC) genetically modified mouse model (23), cultured cells were orthotopically transplanted into the pancreas, which grow in situ to form a tumor characterized by copious neutrophil infiltration (14). Single-cell RNA sequencing (scRNAseq) was performed on total CD11b+CD115−Ly6G+ cells from the bone marrow (BM), spleen, blood, and tumors (n=2, Fig. 1A, fig. S1A and 1B) to characterize neutrophil heterogeneity in the context of their ontogenetic order and origin. Uniform Manifold Approximation and Projection (UMAP) analysis revealed that neutrophils from the bone marrow, spleen, and blood clustered according to their developmental states, from pre-neutrophils (preNeus) to immature neutrophils (IMM 1–2) and mature neutrophils (MAT 1–5) (Fig. 1B, fig. S1C). Integration with a previous dataset containing healthy mouse neutrophils from matching tissues (19) showed that despite potential perturbations from the tumor, no new developmental clusters or trajectories emerged in neutrophils from tissues of the tumor-bearing mice (fig. S1D and E). This thus demonstrated a robust adhesion of neutrophils to a single developmental trajectory within the tissues where they develop and circulate. In contrast, neutrophils acquired unique transcriptional profiles in the tumor (Fig. 1B and fig. S1C) and formed three distinct clusters (T1-T3) separate from the other compartments.
Figure 1. Neutrophils infiltrating the pancreatic tumor undergo further differentiation and converge upon a transcriptionally distinct T3 neutrophil state.
(A) Schematic shows scRNAseq workflow. CD11b+Ly6G+ neutrophils were sorted from the bone marrow, spleen, blood and pancreatic tumor for tumor bearing mice 6 weeks post orthotopic injection (n=2). Each sample was individually tagged with cell-hashing antibodies before they were pooled for analysis with 10X V3 3’ scRNAseq. (see also fig. S1A and Materials and Methods)
(B) UMAP projection of total neutrophils in bone marrow (BM), spleen, blood and tumor show strong enrichment of three clusters (T1, T2 and T3) in the tumor. Louvain clustering was performed and colors correspond to clusters identified.
(C) Low dimensional embedding of all neutrophils using a diffusion-map approach reveals a branch point between mature and tumor neutrophils. Scatterplot shows diffusion components 1 and 2. Light grey arrows are used to indicate the trajectory from precursor, immature and mature neutrophil states. Black arrows denote the branching into T3 neutrophils from T1 and T2 states.
(D) The neutrophil maturation score can be used to identify Louvain clusters along their differentiation trajectory. Histograms show the module score of the maturation gene signature for each cluster identified in (B), with scores closest to 1 being the most mature. Dotted line in grey defines the cut-off for mature neutrophils, and is set at the lower bound of the Mature 2 cluster.
(E) RNA velocity suggests the convergent differentiation of T1 and T2 into T3 neutrophils. RNA velocity vectors are projected on the diffusion map embedding with velocity vectors terminating in the tumor and mature neutrophils.
(F) Principal component analysis of bulk ATAC (Assay for transposase accessible chromatin) sequencing (ATACseq) indicate that changes in chromatin accessibility established in tumor neutrophils cluster them away from other neutrophil subsets. ATACseq was performed for immature (circles) and mature (squares) neutrophils sorted from the bone marrow (grey and dark blue, n= 3 each), spleen (light blue, n=3 each), blood (red, n=3 mature, n=2 immature) and tumor (orange, n =3 each) in wild type (WT) or tumor-bearing (tumor) mice.
(G) Open chromatin regions (OCRs) matched to differentially expressed T3 genes, have increased accessibility only in immature and mature tumor neutrophils. Heatmap shows intensity of fragment mapping across indicated neutrophil subsets, histograms indicate average mapping intensity. T3 genes linked to hypoxia, glycolysis and angiogenesis are annotated. (see also table S2)
(H) Heatmap shows scaled AUC scores of the top 25 transcription factor regulons enriched in T3 neutrophils computed by PySCENIC (see also table S2). Numbers in brackets denote the number of genes assigned to the transcription factor regulon. Transcription factors that also had increased motif enrichment in tumor immature and mature bulk neutrophil populations are in bold.
Next, we sought to uncover links between neutrophil clusters in the tumor and the other tissue compartments by utilizing a diffusion map approach, which orders cells based on transitional probabilities and better preserves differentiation trajectories (24, 25). We found that tumor neutrophils deviated from the normal neutrophil developmental trajectory (Fig. 1C). Specifically, T1 and T2 neutrophils progressed along the tumor-specific branch and converged at the T3 state, suggesting that T1 and T2 may both give rise to the T3 population, which is predicted to be the most terminally differentiated. Because immature and mature neutrophils can infiltrate tumoral tissue (14,15), we investigated the relationship between the maturation and functional states of the T1-T3 neutrophil populations utilizing a neutrophil maturation gene signature (table S1) curated by Xie et al (19), where each cluster in our dataset was graded by a maturation score. T1 neutrophils had a maturation score comparable to immature neutrophils (i.e. IMM 2 cluster), whereas T2 neutrophils had a maturation score comparable to mature neutrophils (MAT 1–5 clusters) (Fig. 1D). Interestingly, RNA velocity (26) indicated a chronological progression from immature neutrophils to T1, and mature neutrophils to T2, suggesting that T1 and T2 neutrophils are transitory states from immature and mature neutrophils that have migrated into the tumor (Fig. 1E). In contrast, T3 neutrophils exhibited a maturation score that was in between T1 and T2 neutrophils (Fig. 1D), representing a potential admixture of T1 and T2 cells. Accordingly, we observed that velocity vectors originating from both T1 and T2 populations terminated in the T3 cluster (Fig. 1E). These findings thus corroborate earlier studies demonstrating that both mature and immature neutrophils infiltrate tumor tissue (4), and become T1 and T2 neutrophils, respectively. More importantly, our data predicts that these stages are transitional, as both T1 and T2 neutrophils remain amenable to further differentiation into T3 neutrophils.
Epigenetic reprogramming of tumor-infiltrating neutrophils
To confirm that T3 neutrophils can be reprogrammed from both immature (T1) and mature (T2) tumor neutrophils, we performed ATACseq (Assay for transposase-accessible chromatin followed by sequencing) on sorted CD101− (immature) and CD101+ (mature) neutrophils from various tissue compartments of control and tumor-bearing mice. Principal component analysis (PCA) revealed that both immature and mature tumor neutrophils clustered separately from neutrophils of other tissues (Fig. 1F), indicating that common changes in chromatin accessibility are imprinted in neutrophils that enter the tumor and are different from those in other tissues, even if they share the same maturation stage. We then identified all open chromatin regions (OCRs) that are differentially accessible in immature and mature tumor neutrophils (table S2), and found increased chromatin accessibility for genes upregulated in T3 neutrophils (T3 genes) in both immature and mature tumor neutrophils (Fig. 1G), including genes involved with hypoxia, glycolysis, and angiogenesis, such as Vegfa and Hk2 (fig. S2A). These changes in chromatin were absent in neutrophils from other tissues, which still shared similar accessibility for canonical neutrophil genes such as Cebpa and Gfi1 (fig. S2B).
Since chromatin accessibility patterns at T3 genes were already altered in immature and mature tumor neutrophils, we investigated the upstream transcription factors of this differentiation program. We utilized pySCENIC (27) to identify transcription factors predicted in T3 neutrophils that had minimal presence in the other neutrophil subsets (Fig. 1H). This was then correlated to motifs found in the bulk ATACseq analysis (fig. S2C), which revealed T3-specific transcription factors with enriched motifs in immature and mature tumor neutrophil OCRs (fig. S2D and E). Among the T3-specific transcription factors, Mafk, Nfe2l2 and Atf3 were implicated in regulating metabolic and oxidative stress typical of the tumor microenvironment (fig. S2D and E). This altered chromatin accessibility landscape in immature and mature tumor-infiltrating neutrophils thus correlated with motif usage of transcription factors governing the T3 state, suggesting that epigenetic changes initiated at the T1 and T2 states are reinforced as they transition towards a T3 state. These findings demonstrate that tumor-infiltrating neutrophils can be reprogrammed by the tumor microenvironment regardless of their stage of maturity by converging transcriptional and chromatin trajectories towards a distinct T3 state.
CD101 and dcTRAIL-R1 discriminate tumor neutrophils states
Having shown that immature T1 and mature T2 neutrophils in the tumor undergo transcriptional and epigenetic reprogramming towards the T3 neutrophils, we next assessed if we could distinguish these subsets by protein expression. This was particularly important, since increases in RNA expression in neutrophils typically precedes protein expression, especially for genes tightly regulated in neutrophil development such as granule proteins.T1, T2 and T3 neutrophils had differential expression of surface marker genes, suggesting that a combination of surface markers could be used to identify them (fig. S3A). Using a multiparametric flow cytometry approach where live CD45+ cells of the PDAC tumor were screened for 249 surface markers, we then clustered all captured cells with InfinityFlow (28, 29), and evaluated co-expression patterns of all tested markers (fig. S3B) which revealed two distinct neutrophil clusters (Fig. 2A). Interestingly, high levels of immune-modulatory/suppressive markers such as dcTRAIL-R1, PD-L1 (CD274) (6), CD14 (13), CD371 (30), VISTA (31), and CD39 (32) distinguished Cluster 2 neutrophils from those of Cluster 1(Fig. 2B). These markers correlated with surface marker genes enriched in T3 neutrophils (fig. S3C), indicating that they may serve to identify the T3 state. Indeed, we found that dcTRAIL-R1 was expressed mainly by a population of tumor neutrophils (Fig. 2C) while having minimal expression in tumor-infiltrating monocytes and macrophages and in neutrophils in the bone marrow, spleen, and blood (Fig. 2C and fig. S3D). In contrast, staining for other markers such as VISTA and CD14 showed expression across multiple neutrophil subsets, and was less restricted to tumor neutrophils (Fig. 4C and fig. S3D). Interestingly, dcTRAIL-R1 expression in tumor-infiltrating neutrophils was also conserved across several other tumor types, including orthotopic breast cancer (median: 20.4%, IQR: 18.0–23.1%) and orthotopic lung cancer (median: 12.1%, IQR: 4.54 – 19.5%) models (fig. S3E). Across all experimental tumor models examined, dcTRAIL-R1 expression was similarly limited to a subset of tumor neutrophils, indicative of a promising candidate marker for T3 neutrophils.
Figure 2. Combinatorial CD101 and dcTRAIL-R1 expression identifies T3, T2 and T1 neutrophils.
(A) UMAP projection of live, CD45+ immune cells within the pancreatic ductal adenocarcinoma tumor reveals two clusters of neutrophils present in the tumor. High parameter flow cytometry was carried on single cell suspensions of pooled tumours (n=10) using the LEGENDScreen™ panel (Biolegend). Live, CD45+ well-compensated FCS were first analyzed and then exported out for analysis using the InfinityFlow package in R. UMAP shows Ly6G expression intensity imputed by InfinityFlow from high (red) to low (blue). Clusters are annotated by canonical surface marker expression. (see also fig. S3B).
(B) Neutrophil cluster 2 has increased expression of immunosuppressive/immunomodulatory surface markers compared to cluster 1. Surface marker expression intensities are shown for curated surface markers from clockwise: dcTRAIL-R1, PD-L1, CD14, CD371, VISTA and CD39. Data are represented as a Z score based on predicted log2 mean fluorescence intensity (MFI) from high (red) to low (blue). (see also fig. S3C)
(C) dcTRAIL-R1 expression marks and is restricted to a separate population of tumor infiltrating neutrophils. Representative contour flow cytometry plots (top) show dcTRAIL-R1 expression against Ly6G expression in the tumor for indicated cell populations. Heatmap (bottom) shows scaled mean fluorescence intensity (MFI) for markers in (B), scaled between 0–1 across all populations, with 0 being the lowest MFI. Populations were analyzed by flow cytometry, where total neutrophils were gated as CD11b+CD115−Ly6G+ in the bone marrow (BM), spleen, blood and tumor. Tumor macrophages (CD11b+Gr-1F4/80hiMHCIIhi) and monocytes (CD11b+Ly6G−Ly6Chi) were gated accordingly. (see also fig. S3D)
(D) Proposed gating strategy to isolate T3, T2 and T1 neutrophils by dcTRAIL-R1 and CD101 expression.
(E) Sorted dcTRAIL-R1+ tumor neutrophils have the highest expression of the T3 transcriptional signature. Heatmap shows scaled Nanostring gene counts (normalized against internal positive controls and housekeeping genes) for T1 (n=4), T2 (n=4) and T3 (n=4) neutrophils sorted according to (D). Genes belonging to either the T3, T2 or T1 transcriptional signature are indicated. (see also fig. S4C).
Figure 4. T3 neutrophils occupy a hypoxic-glycolytic niche in the pancreatic tumor.
(A) Workflow diagram of multiplexed imaging using MICS technology. Cryosections of halved PDAC tumors were placed on slides, fixed and permeabilized, before region of interest (ROI) selection with DAPI staining. Sections were stained for 10 minutes per cycle containing antibodies in FITC, PE and APC. After scanning, sections are photobleached and scanned to subtract background signals. After imaging the desired cycles, images were registered, segmented and exported for conventional flow cytometric annotation of cell types which are further used for spatial statistical analysis with SPIAT.
(B) Tumor pictograph showing ROI1 selection area.
(C) Immunofluorescent image of ROI1 with indicated stain markers of respective tumor regions. Scale bar = 100um.
(D) (left) Expression marker intensity map of CD73 and GLUT1. (right) Co-expression plot of CD71 and GLUT1 marker intensities.
(E) Spatial mapping of each annotated tumor neutrophil subsets in ROI1.
(F) Co-mapping of neutrophil subsets in ROI1.
(G) (left) Gating strategy of CD45-negative, CD105-negative tumor regions demarcated by CD73 and GLUT1. (right) Mapped gated regions on segmented data of ROI1.
(H) Spatial statistical analysis of segmented tumor neutrophils. (left) Average minimum distance of each segmented neutrophil subset to hypoxichighglycolytichigh regions. (right) Co-localisation of tumor neutrophils with hypoxichighglycolytichigh regions, measured using a normalized mixing score for each radius of interaction.
Consistent with its protein expression pattern, gene expression and RNA velocity (indicative of active gene transcription) for the gene encoding dcTRAIL-R1 (Tnfrsf23) were highest in the T3 neutrophil cluster (fig. S4A). Therefore, we attributed the T3 population to the dcTRAIL-R1+ cluster (cluster 2), while the T1 and T2 populations are likely contained within the dcTRAIL-R1− cluster (cluster 1) identified in our InfinityFlow analysis (Fig 2A). Since T1 and T2 neutrophils were transcriptionally identified as immature and mature respectively in our scRNAseq data (Fig. 1D), we next evaluated CD101 expression, which separates immature neutrophils from mature neutrophils (14). Cluster 1 exhibited a two distinct levels of CD101 expression (Fig. S4B), suggesting that CD101 can be used to distinguish T1 and T2 neutrophils within the dcTRAIL-R1− population. Using CD101 and dcTRAIL-R1 to identify the different neutrophil types by flow cytometry (Fig. 2D), we isolated putative T1, T2, and T3 neutrophils and discovered that putative T1 neutrophils possessed a toroidal nuclear morphology resembling immature neutrophils from the bone marrow (Fig. S4D). Putative T2 neutrophils had hyper-segmented nuclei similar to mature BM neutrophils (Fig. S4D) while putative T3 neutrophils had both hyper-segmented and toroidal nuclear morphology (Fig. S4D), congruent with our findings that T3 neutrophils are reprogrammed from both immature and mature neutrophils. Next, we found that dcTRAIL-R1+ neutrophils showed the highest expression of T3 genes (Table S3), whereas dcTRAIL-R1CD101− and dcTRAIL-R1−CD101+ populations were strongly enriched for genes associated with T1 and T2 neutrophils respectively (Fig. 2E and fig. S4C). Finally, all three neutrophil populations were identified in all experimental tumor models examined (fig. S4E), and the frequencies of T1-T3 populations in PDAC as determined by scRNAseq were comparable to the median frequencies of putative T1-T3 populations isolated by flow cytometry (fig. S4E and F). Thus, dcTRAIL-R1 and CD101 expression phenotypically divides tumor neutrophils into three distinct populations and successfully recapitulates the T1-T3 populations defined in our transcriptomic and epigenetic approaches.
Spatial compartmentalization of neutrophils in tumors
Spatial mapping of immune microenvironments in tumors have been highly informative by providing insights into their diversity and functionality based on their localization within the tumor (33). To gain a better understanding of how tumor neutrophils are impacted by their localization in the tumor, we first dissected possible functional differences between the three tumor neutrophil subsets by performing Gene Ontology (GO) analysis on differentially expressed T1, T2, and T3 genes (table S4). T1 neutrophils were enriched for pathways relating to transcription and translation (including genes governing ribosomal biogenesis such as Npm1), and oxidative phosphorylation (proton membrane transport) (Fig. 3A), which are consistent with the immature phenotype of T1 neutrophils (14). T2 neutrophils were enriched in pathways related to transcriptional regulation, amide and reactive oxygen species metabolism, and immune responses as well as type I interferon genes including Ifit1, Ifit2, Ifit3, and Isg15 (Fig. 3B), which likely reflect neutrophil activation upon tumor infiltration. Finally, T3 neutrophils featured pathways of cell stress and survival, including response to hypoxia, oxidative stress, and glycolysis, (Fig. 3C), suggesting adaptation to the hostile tumor environment. Interestingly, T3 neutrophils were also enriched for angiogenic genes, including Vegfa, Thbs1 (34), and Lgals3 (35) that combined, were suggestive of a strong pro-tumoral role.
Figure 3. Spatial compartmentalization of T1, T2 and T3 neutrophils in the pancreatic tumor.
(A) Chord diagram shows differentially expressed genes (DEGs) in T1 neutrophils that are enriched for gene ontology pathways linked to transcription and oxidative phosphorylation.
(B) Chord diagram shows DEGs in T2 neutrophils that are enriched for gene ontology pathways linked to metabolism and immune response.
(C) Chord diagram shows DEGs in T3 neutrophils that are enriched for gene ontology pathways linked to survival and angiogenesis. (A-C) Bars associated with each gene are colored by strength of fold change of differential expression, and are sized based on the number of pathways it interacts with. (see also table S4 for DEG lists)
(D) Spatial transcriptomic analysis workflow. PDAC tumors were isolated and cut into quarters, where the sharp edges denote the core facing regions, before flash-freezing. Fresh frozen PDAC tumors were sectioned and placed on 10X Visium slides containing spatially barcoded capture spots. After processing and sequencing, the data was clustered spatially (BayesSpace) and cell type deconvolution was performed (Cell2location). Gene signatures of various biological processes were then probed and mapped with the UCell package.
(E) Tumor neutrophils localize to different spatial clusters. Projection of T3, T2 and T2 neutrophil enriched spots identified by Cell2location on merged UMAP derived from BayeSpace enhanced clustering analysis of tumor sections (n=4)
(F) Merged UMAP representation of spots of tumor sections analysed, color-coded according to BayesSpace identified clusters (top). Violin plots show frequency of T1, T2 and T3 neutrophils enriched spots mapping to each cluster (bottom).
(G) Spatial mapping of T1, T2 and T3 neutrophils across tumor sections (n=4) by Cell2location. Black lines denote the outline of the section, grey colored areas indicated the excluded DAPI−panCKhi regions annotated to be fibrotic/necrotic. Spots are filtered based on Ly6G positive staining (see also fig. S6B).
(H) Quantification of deconvoluted T3 neutrophil-enriched spots falling into high or low scoring spots for GO ontology pathways: glycolytic (GO:0061621), hypoxic (GO:001666), and angiogenic (GO:0045766). Boxplots show median (centre line) with standard deviation (whiskers). p<0.05*, by one-tailed T test.
We next characterized how neutrophils with distinct transcriptional states would interact with the neighboring tumor microenvironment. Using DAPI and pan-cytokeratin (panCK) co-staining, we noted that our orthotopic tumors were mainly comprised of tumor cell regions (PanCKhighDAPIhigh), fibrotic/necrotic regions (panCKhighDAPI−/low), and sparse stromal regions along the edges (panCK−/lowDAPIhigh) (fig. S5A and B). We performed spatial transcriptomics on four cryo-sections of three different PDAC tumor samples, taking two cryosections from one tumor to evaluate variability within the same tumor (Fig. 3D). As expected, necrotic regions with low numbers of DAPI+ cells contained low quality counts, which were manually annotated and filtered to mitigate RNA contamination from nearby spots and improve downstream analyses (fig. S5C and D). We next assessed the tumor architecture by utilizing BayesSpace (36) to identify transcriptionally similar neighborhoods within the tumor (fig. S5E). Indicative of the complexity of the tumor environment, the two cryosections (region of interest, ROI1, ROI2) taken from different regions of the tumor showed different distributions of spatial clusters identified (fig. S5E). Nevertheless, clustering and UMAP embedding revealed 10 clusters shared across all four sections (fig. S5E), each possessing specific functional pathways associated with their distribution (fig. S5F). For example, regions enriched for pathways related to epithelial-to-mesenchymal transition (EMT, clusters 1 and 10), typically associated with the invasive tumor front (37), were present closer to the tumor periphery, while regions associated with hypoxia (cluster 6) and cell cycle progression (G2-M checkpoint, cluster 2) were located close to the core of the tumor (fig. S5E and F).
Utilizing Cell2location (38) with a pre-annotated scRNAseq dataset (fig. S6A) allowed us map the full transcriptional signature of each cell cluster within the tumor and estimate cell type abundances within each spot. To ensure the accuracy of neutrophil assigned spots, we ascertained the presence of Ly6G immunofluorescence staining falling within the spot as a selection cutoff (fig. S6B). Cell2location revealed that T1, T2 and T3 neutrophils mapped to different regions within the tumor (Fig. 3E), with visualization of T3 enriched spots on the UMAP embedding indicating that T3 neutrophils had the highest enrichment in spatial cluster 6, which was associated with hypoxia (Fig. 3F). T3 neutrophils were also observed to be mostly distributed adjacent to PanCKhighDAPI−/low necrotic zones, (Fig. 3G), consistent with their localization to cluster 6 (fig. S5F). In contrast, T1 neutrophils were highly enriched in in cluster 1 (EMT), 7 and 8 (IL-2 and STAT5 signaling) regions, while T2 neutrophils were mostly found in cluster 3 associated with p53 signaling (Fig. 3F). Both T1 and T2 annotated spots were generally localized closer to the tumor periphery compared to T3 neutrophils (Fig. 3G). Our data thus reveals that neutrophils are spatially organized within the tumor, and T3 neutrophils occupy a tumor niche that is distinct from that of T1 or T2 neutrophils.
Reprogrammed neutrophils occupy a hypoxic and glycolytic tumor niche
Since T3, but not T1 or T2, neutrophils had transcriptional and epigenetic profiles that were enriched for hypoxia, glycolysis and angiogenesis pathways, we evaluated whether this was reflected in their localization to hypoxic, glycolytic or angiogenic regions within the tumor. All spots within the tumor region were first scored for GO pathways corresponding to glycolysis (fig. S6C), hypoxia (fig. S6D) and angiogenesis (fig. S6E), with a 50th percentile cut-off employed to determine low and high regions. We then determined the frequency of neutrophil-containing spots falling into low and high regions for each tumor neutrophil subset. Notably, T3 neutrophils were found at greater frequencies in high scoring regions for glycolysis (68.8% ± 19.7), hypoxia (53.3% ± 4.65) and angiogenesis (62.2% ±14.2) (Fig. 3H). In contrast, the majority of T2 (fig. S6F) and T1 (fig. S6G) neutrophil-containing spots fell into regions with low glycolysis, hypoxia and angiogenesis scores, indicating that partitioning of the neutrophil subsets potentially results from T3 neutrophils occupying a specific regional hypoxic and glycolytic tumor niche.
We next set out to validate these observations at single-cell resolution using MACSima imaging cyclic staining (MICS) (Fig. 4A), where iterative immunofluorescence staining and photobleaching allows for evaluation of large numbers of antibody targets within the same tissue slice (39). Optimization was first carried out to determine working markers within orthotopic pancreatic tumor tissues (fig. S7A and B), given that MICS staining times were relatively short at 10 minutes. Testing revealed that CD29, Galectin-3 and CD44, which typically mark fibroblasts (40–42) displayed the greatest staining at peripheral stromal/desmoplastic (panCK−/lowDAPIhi) regions (fig. S7C), while CD31 and CD105 marked vessels within the stromal and tumor region (panCK+). Notably, CD105 was able to better resolve intra-tumoral vessels (fig. S7C), consistent with their role in marking tumor-associated endothelium (43). To identify hypoxic regions within the tumor, Hif1a, CD39 and CD73 staining was evaluated. CD73, an adenosine monophosphate ectoenzyme, has been found to be directly induced by hypoxia and Hif1a in cancer (44–47). Immunofluorescence analysis revealed diverse staining patterns of CD73 in both stromal/desmoplastic and tumor (panCK+) regions (fig. S7C), resembling the spatial distribution of hypoxia gene signature scores observed in our 10x Visium analysis (fig. S6B). In contrast, Hif1a did not stain within the MICS timeframe, while CD39 mostly identified vessels (fig. S7C). Similarly, GLUT1 (glucose transporter 1) staining showed regional restriction (fig. S7C) similar to glycolysis signature scores (fig. S6B). Thus, CD73 and GLUT1 representative staining was utilized to define the hypoxic and glycolytic tumor niche. T1, T2 and T3 neutrophils were identified by staining for Ly6G, CD101 and dcTRAIL-R1 based on our flow cytometric strategy (Figure 2D) and successfully annotated in both confocal (fig. S7D) and MICS (fig. S7E).
We carried out MICS for five different ROIs across two different orthotopic pancreatic tumors at 4–6 weeks post injection. ROIs were selected to unequivocally cover an entire region from the left to right margin encompassing the core of the tumor (fig. S8A–B), and we observed staining across the full panel for all markers surveyed (fig. S9). We next specifically evaluated staining for the tumor vasculature (CD105), hypoxic (CD73) and glycolytic (GLUT1) across all five ROIs (Fig. 4C and fig. S8A–B). CD105+ blood vessels were present as a diffuse network scattered throughout the tumor (Fig. 4C and fig. S8A–B). A gradient of CD73 staining was apparent throughout the tumor, while GLUT1 staining was more tightly localized, however GLUT1hi areas strongly co-localized with CD73hi regions marking a distinct hypoxic and glycolytic niche (Fig. 4D and fig. S8A–B). Annotation of T1 (Ly6G+CD101−dcTRAIL-R1−) and T2 (Ly6G+CD101+dcTRAIL-R1−) neutrophils (fig. S8C) revealed that T1 and T2 neutrophils were distributed throughout the tumor parenchyma in two different patterns reflective of their likely routes into the tumor – bordering the tumor edges in CD73− stromal regions or diffused throughout GLUT1−CD73+ regions, both of which contained a substantial network of CD105+ vessels (Fig. 4E and fig. S8A–B). In contrast, T3 (Ly6G+CD101−/+dcTRAIL-R1+) neutrophils across all ROIs were more likely to cluster together at the CD73hiGLUT1hi hypoxic and glycolytic niche (Fig. 4E), thus giving rise to a clear spatial segregation of the three neutrophil populations as suggested by our 10x Visium analysis (Fig. 4F and fig. S8A–B).
Next, we agnostically quantified the three neutrophil subgroups’ relative locations within the tumor. Using marker intensities on segmented cells, we excluded CD45+ immune and CD105+ endothelial cells, and subdivided the remaining cells into three distinct regions – stromal (CD73GLUT1−), tumor parenchyma (CD73+GLUT1−) and the hypoxichighglycolytichigh tumor niche (CD73hiGLUT1hi) (Fig. 4G and fig. S8C). In all five ROIs, T3 neutrophils had the shortest average minimum distance to the hypoxichighglycolytichigh niche compared to T1 and T2 neutrophils (Fig. 4H). We then assessed normalized mixing scores, which quantify proportion of direct touches between the target regions and reference neutrophil cells (target-reference) against reference-reference interactions (Feng et al., 2023). T3 neutrophils had the greatest mixing scores within hypoxichighglycolytichigh tumor regions, which was conserved with each radius of area investigated (Fig. 4H). Scores for the stromal region showed a trend for highest scores in T1, followed by T2, with T3 being minimally detected within the stroma (fig. S8D). Finally, T2 neutrophils trended towards higher proportions of interspersion within the tumor parenchyma compared to T1 and T3 neutrophils (fig. S8D). These observations suggest a possible migration pattern of neutrophils from vessels into tumor stroma/parenchyma as T1 and T2 neutrophils. Although the temporal timeline for when T3 reprogramming occurs is still unclear, our spatial data implies that T3 neutrophils likely migrate into and occupy the specialized hypoxic-glycolytic tumor niche. This is consistent with our finding that transcription factor regulons enriched in T3 neutrophils (Fig. 1H) include those upregulated in response to hypoxia (Hif1a, Bhlhe40) and to metabolic and ER stress (Atf4, Ddit3, Atf3, Nfe2l2). As such, epigenetic and transcriptional upregulation of these pathways in T3 neutrophils could prime them for added survival and/or function within these microenvironments. Collectively, our data provides a spatial representation by which tumor neutrophils converge upon the occupancy of a specialized tumor niche upon reprogramming.
Deterministic reprogramming of neutrophils in the tumor
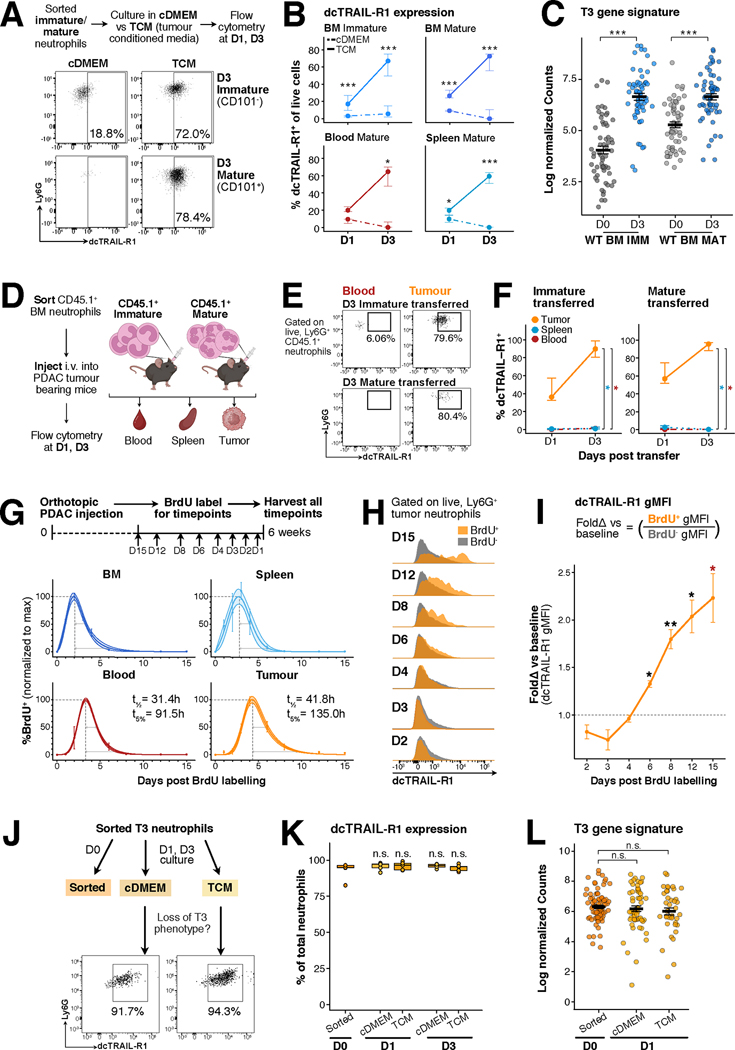
Because both mature and immature neutrophils appear to functionally converge inside the tumor, we examined the impact of the maturation state on the acquisition of the T3 signature. We first isolated immature and mature neutrophils from wild-type mice and cultured them with tumor conditioned media (TCM), a strategy that allowed us to mimic the infiltration of immature and mature neutrophils into the tumor microenvironment (Fig. 5A). We then measured upregulation of dcTRAIL-R1 on cultured neutrophils across multiple timepoints as a proxy to estimate the acquisition of the T3 profile (Fig. 5A). After three days, a substantial proportion of bone marrow immature and mature neutrophils cultured in TCM were dcTRAIL-R1+ but not those cultured in control media (cDMEM) (Fig. 5A). We obtained similar results regardless of tissue origin or maturation state, such that neutrophils isolated from the bone marrow, spleen and circulation cultured in TCM all exhibited dcTRAIL-R1 upregulation and concurrent increased survival (Fig. 5B, fig. S10A–B). Specifically, exposure to TCM triggered dcTRAIL-R1 expression 24h hours into culture, prolonged survival up to three days in comparison to culture in cDMEM (fig. S10A–B) and induced the upregulation of the T3 gene signature in immature and mature neutrophils (Fig. 5C). We then further assessed if culture in hypoxic conditions would enhance neutrophil survival or dcTRAIL-R1 upregulation of wild type neutrophils. Culture in hypoxic conditions slightly augmented neutrophil survival in vitro, especially for mature neutrophils (fig. S10C), but did not induce dcTRAIL-R1 expression (fig. S10D), indicating that hypoxic conditions are not the main driver of T3 reprogramming. Adoptive transfer of CD45.1+ immature and mature neutrophils into tumor-bearing mice (Fig. 5D) further confirmed that exposure to tumor microenvironment was necessary for their upregulation of dcTRAIL-R1 (Fig. 5E), and dcTRAIL-R1 upregulation in vivo followed the same kinetics as in vitro (Fig. 5F, fig. S11A). Hence, direct contact with tumor-derived stimuli is sufficient to extend the survival of wild-type neutrophils and acquisition of the T3 phenotype.
Figure 5. Reprogramming within the tumor environment results in long-lived, terminally differentiated T3 neutrophils.
(A) Experimental set-up of in vitro culture of sorted neutrophils from wild type (WT) mice in control media (complete DMEM, cDMEM) versus tumor conditioned media (TCM). Representative flow cytometry plots show dcTRAIL-R1 expression increases on sorted immature and mature WT neutrophils from the bone marrow (BM) after three days of culture in TCM but not in cDMEM.
(B) Neutrophils cultured in TCM upregulate dcTRAIL-R1 over time. Lineplots show the percentage of dcTRAIL-R1+ cells gated as in (B), dots indicate the median, error bars indicate Q1 and Q3 intervals for neutrophil subsets cultured in cDMEM (dotted line) and TCM (solid line) over 1 and 3 days. Each group contains the following number of samples – Day 1 cDMEM: BM immature (n=8), BM mature (n=8), Spleen Mature (8), Blood Mature (3), Day 1 TCM: BM immature (n=8), BM mature (n=8), Spleen Mature (8), Blood Mature (5), Day 3 cDMEM: BM immature (n=10), BM mature (n=10), Spleen Mature (10), Blood Mature (4), Day 3 TCM: BM immature (n=10), BM mature (n=10), Spleen Mature (10), Blood Mature (5). p<0.05*, p<0.01**, p<0.001***, by Mann-Whitney U test.
(C) Neutrophils cultured in TCM upregulate the T3 gene signature. Scatter dot plots for T3 gene signature expression in BM immature (WT BM IMM) and mature (WT BM MAT) that were freshly sorted (D0) or cultured for 3 days (D3), n=3 for all samples. Each dot denotes a single gene, lines denote the mean, error bars show standard error mean (SEM). p<0.001***, by Wilcoxon signed-rank test with Bonferonni’s correction, comparisons indicated on graph.
(D) Experimental set-up for transfer of CD45.1+ neutrophils into pancreatic tumor bearing mice. Sorted WTBM immature and mature neutrophils were intravenously injected into WT PDAC tumor-bearing mice. At 1- and 3-days post transfer, CD45.1+ neutrophils were evaluated within the blood, spleen and tumour for dcTRAIL-R1 expression by flow cytometry.
(E) Upregulation of dcTRAIL-R1 expression is restricted to the tumor. Representative flow plots show dcTRAIL-R1 expression present on transferred CD45.1+ immature and mature neutrophils present in the blood or tumor at 3-days post transfer.
(F) Lineplots show proportion of WT BM CD45.1+ immature (n=4 at day 1, n=3 at day 3) or mature neutrophils (n=3 at both timepoints) expressing dcTRAIL-R1. Each dot denotes a single gene, lines denote the mean, error bars show standard error mean (SEM). p<0.05*, by Kruskal-Wallis test with Dunn’s post-test.
(G) Experimental set-up for BrdU pulse-labelling in tumor-bearing mice. WT mice received orthotopic injection of the pancreatic ductal adenocarcinoma (PDAC) cells, and the tumor was allowed to grow. At days 15 (n=4), 12 (n=4), 8 (n=5), 6 (n=3), 4 (n=4), 3 (n=4), 2 (n=3), and 1 (n=4) prior to the harvest, mice were injected with BrdU, thus labelling proliferating neutrophils within the bone marrow and spleen. At day 42 (6 weeks) post injection, mice were harvested, and BrdU+ neutrophils within the bone marrow, spleen, blood and tumor were quantified. BrdU percentages at all timepoints were then normalized to the maximal BrdU+ percentage value for each tissue, which was set to 100%. Dots shows mean expression with error bars denoting 95% confidence intervals. A mathematical model capturing the full temporal window was fitted to estimate half-life (t1/2)and lifespan (t5%)for each organ, denoted on each plot in hours (see also Methods and fig. S12C).
(H) BrdU labelled neutrophils upregulate dcTRAIL-R1 over time. Histograms show geometric mean fluorescence intensity (gMFI) of dcTRAIL-R1 within BrdU+ (orange) and BrdU− (grey) neutrophils over day 2,3,4,6,8,12 and 15 timepoints post BrdU labelling.
(I) Quantification of gMFI in (H). Line plots fold change of dcTRAIL-R1 gMFI of BrdU+ against BrdU− neutrophils. BrdU− neutrophils served as a measure of baseline dcTRAIL-R1 gMFI within the tumour. Each dot denotes the mean, with error bars showing SEM. p<0.05*, p<0.01**, by Mann-Whitney U test, one-tailed, alternative = “greater”.
(J) Experimental set-up of in vitro culture of sorted T3 neutrophils from PDAC mice in cDMEM or TCM at 1 or 3 days. Representative flow cytometry plots show dcTRAIL-R1 expression is retained on T3 neutrophils after 1 day of culture in both cDMEM and TCM.
(K) Boxplots show quantification of frequency of dcTRAIL-R1+ neutrophils (n=5) in (I), where lines denote the mean. p = n.s. (non significant), by Kruskal-Wallis followed by many-to-one U test comparing against the D0 timepoint.
(L) T3 neutrophils cultured overnight do not downregulate the T3 gene signature. Scatter dot plots for T3 gene signatures in sorted, cDMEM or TCM cultured neutrophils (n=3 each), each dot denotes a single gene, lines denote the mean, error bars show standard error mean. p = n.s., by Kruskal-Wallis followed by Dunn’s post-test.
Both TCM-cultured immature and mature neutrophils upregulated dcTRAIL-R1 expression and the T3 gene signature to an equal degree, indicating that immature neutrophils could directly be reprogrammed into T3 neutrophils (Fig. 5A–C). RNA velocity analysis suggested that this occurs in a stepwise fashion as immature neutrophils transit into T1 neutrophils, and subsequently differentiate into T3 neutrophils (Fig. 1E). As expected, transfer of CD45.1+ immature neutrophils resulted in the appearance of dcTRAIL-R1−CD101− T1 neutrophils within the tumor one day posttransfer (fig. S11B). In contrast, only dcTRAIL-R1−CD101+ T2 neutrophils, and not T1 neutrophils, were observed in the tumor with CD45.1+ mature neutrophil adoptive transfer (fig. S11B). Both T1 and T2 populations derived from transferred cells were undetected after three days, as the majority of these transferred cells completed their reprogramming into dcTRAIL-R1+ T3 neutrophils (fig. S11C). We further corroborated the transitory nature of T1 and T2 subsets by in vitro culture (fig. S11D–E). Isolated T1 and T2 neutrophils upregulated dcTRAIL-R1 expression after 24 hours in culture. Unlike tumor-naïve neutrophils, dcTRAIL-R1 upregulation was independent of the media utilized for both T1 (fig. S11F) and T2 neutrophils (fig. S11G). As such, T1 and T2 neutrophils reflect immature and mature neutrophil populations captured in the process of T3 differentiation, and thus do not require any further input from tumor soluble factors. Taken together, our findings uncover a deterministic program within neutrophils that enables them to acquire the T3 phenotype independent of their maturation stage.
T3 neutrophils are long-lived, terminal effectors within the tumor microenvironment
To better understand the temporal regulation of neutrophil differentiation into the T3 state within the tumor microenvironment in vivo, we administered BrdU intravenously at selected timepoints prior to harvest, pulse-labelling all proliferating neutrophil precursors (Fig. 5G). This approach defines a timestamp at the point of labelling, allowing us to evaluate all timepoints simultaneously, thereby minimizing batch effects. Modelling the disappearance rate from the peak of BrdU+ signal (fig. S12A–B) and incorporating the entire temporal interval measured allowed the prediction the neutrophil half-life and lifespans (fig. S12C), reflecting their dwell time within each tissue compartment (Fig. 5G).
The earliest peak of BrdU neutrophils was observed in the bone marrow and spleen, due to active granulopoiesis within these organs in the tumor-bearing state, while recruitment of BrdU-labelled neutrophils into the blood and subsequently the tumor placed the peak at roughly 4–5 days post labelling (Fig. 5G). As the dwell times of neutrophils in the bone marrow and spleen are complicated by continual neutrophil production, we focused our comparison between the blood and tumor neutrophils. Blood neutrophils had a half-life of 31.4 hours, reflecting an increased transit time in circulation compared to previously predicted times for wild type blood neutrophils (48). Importantly, tumor neutrophils had an even longer half-life (41.8h) and a predicted lifespan of 135 hours (up to 5.625 days) (Fig. 5G, fig. S12C–D), representing a remarkable extension of neutrophil lifespan within the tumor microenvironment. Notably, when compared to the unmarked BrdU− fraction, the proportion of BrdU+ dcTRAIL-R1+ neutrophils increased over time, surpassing baseline levels at day 6 post labelling (fig. S12E and F). Newly recruited BrdU+ tumor neutrophils already expressed dcTRAIL-R1 at 1-day post-labelling (fig. S12E), suggesting that T3 reprogramming is initiated upon tumor entry. dcTRAIL-R1 expression steadily increased and was the highest in neutrophils at 15 days post-labelling, (Fig 5H and I), confirming that acquisition of the T3 phenotype was associated with their extended lifespans in vivo. Our results show that a notable increase in the duration of neutrophil half-life and residence in the tumor, as compared to those in non-tumor tissues (fig. S12D) – indicating that neutrophils can survive long enough within the tumor to undergo reprogramming and sustained persistence within the tumor as long-lived dcTRAIL-R1+ T3 neutrophils.
To examine if the T3 state is transitory or is stably maintained once it has been acquired, we isolated and cultured T3 neutrophils overnight, and up to 3 days in the presence or absence of TCM (Fig. 5J). Survival of T3 neutrophils in culture was not affected by the media type (fig. S12G), and dcTRAIL-R1 expression was maintained despite the absence of tumor-derived factors (Fig. 5K) with expression levels comparable to freshly isolated T3 neutrophils at both timepoints (fig. S12H). Additionally, T3 neutrophils did not downregulate their gene signature in culture (Fig. 5L). This in vitro evidence indicates that once neutrophils have been reprogrammed to T3 neutrophils, they do not revert their phenotype in the absence of supportive tumor factors and that they represent the terminally differentiated neutrophil population within the tumor.
T3 neutrophils are pro-angiogenic and promote tumor growth
Prior studies have identified key roles for neutrophils in tumor progression, especially via the promotion of tumor angiogenesis (49–51). We therefore sought to determine the functional specialization of T1-T3 neutrophils in promoting tumor growth. Given their phenotypic stability, distribution near angiogenic regions, and the expression of a pro-angiogenic transcriptional signature (Fig. 3C), we speculated that T3 neutrophils promoted angiogenesis from their hypoxic-glycolytic niche to support continual tumor growth. In contrast, we anticipated that the transient T1 and T2 neutrophil states would not yet feature pro-angiogenic ability. T3 neutrophils had the highest transcript (Fig. 6A) and protein expression (Fig. 6B–C) of Vegfa (vascular endothelial growth factor a) in comparison to the other neutrophil subsets in the tumor and periphery. We evaluated whether T3 neutrophils had a greater capacity to induce blood vessel formation in vivo using a modified Matrigel plug assay and measured angiogenesis by the rate of vascular flow measured by Doppler imaging (48, 52) (fig. S13A). Matrigel plugs co-injected with T3 neutrophils showed the greatest flux intensity when compared to control WT BM neutrophil plugs within the same mouse (fig. S13B) and this increase in vascularization, while modest, was consistent across all mice observed in the assay (fig. S13C). In contrast, co-injection of T2 and T1 neutrophils did not enhance vascular flow (fig. S13B–C), indicating that T3 population contains the most potent angiogenic ability (Fig. 3C).
Figure 6. Pro-tumoral T3 neutrophils promote tumor growth and associate with poorer patient outcomes.
(A) T3 neutrophils have the highest expression of Vegfa transcripts. UMAP projection of total neutrophils in bone marrow (BM), spleen, blood and tumor shows expression density of Vegfa.
(B) T3 neutrophils have the highest expression of VEGFα. Representative flow plots show intracellular VEGFα protein staining in neutrophils from the spleen and T1, T2 and T3 neutrophils in the pancreatic tumor.
(C) Boxplots quantify VEGFα expression as shown in (B). Boxplots show median (centre line) with standard deviation (whiskers). p<0.05*, p<0.01**, by Kruskal-Wallist followed by many-to-one U test comparing against T3 neutrophils.
(D) T3 neutrophils promote rapid tumor growth. Schematic of experimental set-up to determine the ability of neutrophils to promote tumor growth in vivo. Equal numbers of neutrophils and pancreatic ductal adenocarcinoma cells (PDAC) were mixed in Matrigel before subcutaneous injection into the right flank. Neutrophils in the tumor was boosted on day 3 (D3) and D7, by direct subcutaneous injection into the visible matrigel plug/tumor. Tumor growth was measured by calipers weekly until the day 28 endpoint. Line plots show volume of measured subcutaneous tumors co-injected with PBS (n=8), WT BM immature (WTBM IMM, n = 10), WT BM mature (WTBM MAT, n =12), T1 (n = 7), T2 (n=12), and T3 (n = 7) neutrophils. Dots show mean with error bars indicating standard error of mean (SEM), over all three measurement timepoints. p<0.05*, p<0.01**, by Kruskal-Wallis test followed by many-to-one U test comparing all other conditions to PBS co-injection.
(E) Quantification of PDAC subcutaneous tumor incidence after neutrophil co-injection (see fig. S13D for quantification method). Piecharts show frequency of mice with tumours (light grey) or no tumor growth (dark grey) at day 28 endpoint in (A).
(F) Tumors co-injected with T3 neutrophils had the biggest weights compared other conditions. Each dot represents one biological replicate, with boxplots showing median with interquartile range, and whiskers indicating lowest and highest measurement. p<0.05* by Kruskal-Wallis test followed by many-to-one U test comparing all other conditions to PBS co-injection.
(G) Antibody neutralization of VEGFα reduces T3-mediated acceleration of tumor growth. Schematic of experimental set-up with sorted T3 neutrophils are co-injected with PDAC tumor with anti-VEGFα targeting antibody (n=10) or isotype control (n=7). WTBM mature neutrophils were utilized as controls (anti-VEGFα targeting antibody, n=4, isotype, n=5). Neutrophils were further injected on D3 and D7 with the corresponding antibody added, and tumour growth was measured at D14 and D21, 7 and 14 days post tumor injection. Boxplots show median tumour volume with whiskers representing the interquartile range. p<0.05* by by Mann-Whitney U test.
(H) Visualization of CD31 vessels within T3 and WTBM MAT co-injected subcutaneous tumours. Subcutaneous tumours from (A) for T3 (n=3) and WTBM MAT (n-3) were dissected, optically cleared and permeabilized, stained with anti-CD31 antibody, and imaged in 3D at 0.65X (100% of total volume) and 2X resolution (35% of total volume starting from midpoint). Representative 3D immunofluorescence images show T3 (top, quarter) and WTBM MAT (bottom, whole) co-injected tumors, with tumour margins marked out in white dotted lines and indicated.
(I) Subcutaneous PDAC tumors co-injected with T3 neutrophils have greater CD31 vessel density within the tumor core. Quantification of CD31 staining intensity at 2X resolution as in (E). CD31 staining was surfaced with a seedpoint of 16.2, and binned in 10% quantiles according to the distance from left or right margins towards the tumour core. To account for differences in tumor sizes, CD31 staining intensity was further normalized over total slice volume for each quantile. Lineplots represent volume-normalized staining intensity, with dots representing the mean and errors bars indicating SEM for T3 (orange) or WTBM MAT (grey) co-injected tumours. p<0.05* by Mann-Whitney U test, one-tailed, alternative = “greater”.
(J) Antibody-mediated blockade of T3 neutrophils reduces their ability to promote rapid tumour growth. Schematic of experimental set-up - sorted T3 neutrophils are co-injected with PDAC tumor with anti-dcTRAIL-R1 targeting antibody (n=8) or isotype control (n=8). T3 neutrophils were further injected on D3 and D7 with the corresponding antibody added, and tumour growth was measured at D14 and D21, 7 and 14 days post tumor injection. Boxplots show median tumour volume with whiskers representing the interquartile range. p<0.05* by by Mann-Whitney U test.
(K) The T3 neutrophil gene signature is associated with poorer patient overall survival (OS) and disease-free survival (DFS) in pancreatic cancer. Kaplan-Meier plots show OS (top) and DFS (bottom) for patients in the TCGA-PAAD (The Cancer Genomics Atlas, Pancreatic Adenocarcinoma) and PACA-AU (international Cancer Genome Consortium Pancreatic Cancer-Australia). Patients were split into high and low expression of the T3 curated signature. Median OS and DFS survival are represented on the graph when available. Events are represented by vertical lines and were defined from days to death (from initial pathological diagnosis) or days to first event (from initial treatment, for TCGA, and from clinical disease-free diagnosis for PACA-AU)). p<0.05* as calculated by log-rank test.
(L) The T3 neutrophil gene signature is associated with poorer patient overall survival in a subset of solid tumor human cancers. Each data set within the curated TCGA Pan Cancer data set was scored for T3, T2 and T1 signatures (see also table S4). Forest plots show hazard ratio (HR) scores associated with patient overall survival (OS) that were significant (p<0.05) by Cox proportional hazards test across all signatures. Dots indicate the calculated HR, whiskers indicate 95% confidence intervals.
The tumor triggers angiogenesis in order to maintain the supply of oxygen and other nutrients to the core of the tumor to sustain continual growth (53). To test whether T3 neutrophils residing in their hypoxic-glycolytic niche at the tumor core support this angiogenic switch, we modified our PDAC model by implanting PDAC tumors subcutaneously to enable longitudinal tumor size measurements. We then assessed whether injection of neutrophils within the tumor affected their subsequent growth (Fig. 6D). PDAC cells co-injected with T3 neutrophils formed tumors that grew rapidly (Fig. 6D), while co-injection with other neutrophil types had little influence in tumor growth. T3 co-injected had a 100% tumor growth rate (Fig. 6E, as evaluated in fig. S13D), and had the greatest mass at the endpoint (Fig. 6F). Interestingly, we noticed that T2-coinjected tumors showed the lowest tumor incidence rates (5/12) (Fig. 6F), suggestive of increased tumor rejection, consistent with the expression of pro-inflammatory genes in T2 neutrophils detected in our scRNAseq dataset (Fig. 3B). T3-coinjected tumors continued to grow rapidly for up to 3 weeks past the last injection of neutrophils, suggesting that even after their disappearance T3 neutrophils generated long-lasting effects that sustained this accelerated growth rate. Neutralization of VEGFα in our model resulted in the reduction of growth in T3 co-injected tumors, but had no impact on the growth of WTBM MAT co-injected tumors (Fig. 6G). To assess if this growth inhibition was due to increased angiogenesis driven by T3 neutrophils, we optically cleared T3- and WTBM MAT co-injected tumors (fig. S13E), and visualized the intratumoral 3D vessel network via CD31 staining (Fig. 6H). Notably, T3 tumors had greater CD31 staining density towards the tumor core (Fig. 6H, Movie S1), while the vessel network in WTBM MAT co-injected controls showed greater distribution along the tumor edges (Fig. 6H, Movie S2). Normalization for tumor size revealed that both T3 and WTBM MAT co-injected tumors had similar densities of CD31+ blood vessels (fig. S13F) but drastically differed in their distribution (Fig. 6I). Although T3 neutrophils were the highest expressors of Vegfa in the pancreatic tumor, Vegfa expression was also detected in macrophage populations (fig. S13G), indicating that neutrophils may not be the sole pro-angiogenic contributor for tumor growth support. Nonetheless, blockade of T3 neutrophils with an anti-dcTRAIL-R1 antibody decreased tumor growth in T3 co-injected tumors compared to isotype controls (Fig. 6J), indicating that T3 neutrophils are the predominant cells responsible for increasing the tumor growth rate, most likely through vascular remodeling.
Evidence for conserved neutrophil reprogramming across tumor types and human PDAC
Phenotypic differences in tumor-infiltrating neutrophils have been observed across mouse and human cancers, and multiple subsets have been characterized by differential surface marker or transcriptome expression (6–13). Whether such neutrophil profiles are conserved across tumor types and species remains unclear. We therefore assessed if T1-T3 neutrophil states can be detected in previously published mouse cancer scRNAseq datasets containing annotated neutrophils (11, 13). We projected these datasets onto a reference UMAP embedding, mapping each cell on the same UMAP space as our dataset (fig. S14A). As in the PDAC model, neutrophils within Lewis lung carcinoma (LLC) tumors (13) could be assigned to the T1-T3 states, while spleens from wild type or tumor-bearing mice mostly contained non-tumor neutrophil clusters (fig. S14B). An intermediate neutrophil subset identified in cancer-bearing individuals (PMN2 in (13)) was enriched in the preNeu and IMM 1–2 clusters (fig. S14C), while T1-T3 clusters scored highly for the tumor-specific neutrophils (PMN3; fig. S14D). Similarly, re-analysis of a different study showed that neutrophils from KP.10 tumor-bearing lung model (11) mapped as T1-T3 clusters, while the normal lung tissue was mostly enriched for mature neutrophils (fig. S14E). Likewise, neutrophil types identified as tumor-specific neutrophil clusters in this model (mN3–6 in (11)), corresponded to T1-T3 clusters (fig. S14F), confirming that the tumor microenvironment induces a prototypical transcriptional trajectory that is deterministic in nature. In contrast, the signature of neutrophils from healthy lungs (48) was largely independent of tumor-induced transcriptional changes (fig. S14G). Taken together, these data suggest that reprogramming of tumor neutrophils is conserved across different tumor types, and that T3 neutrophils represent terminal differentiated tumor neutrophils.
To examine cross-species conservation of the neutrophil tumoral trajectory found in mice, we evaluated if our neutrophil classification could account for heterogeneity present in human tumors. We examined independent scRNAseq datasets (54, 55) from two human PDAC cohorts (fig. S14H and I) and mapped them to a simplified reference UMAP embedding via label transfer (fig. S14J). T3 and T2 neutrophils were amply labelled in both datasets, and a smaller cluster of T1 neutrophils was also identified (fig. S14K and L). These tumor neutrophil subsets were predominantly enriched in the pancreatic tumor and not in adjacent normal pancreatic tissue (fig. S14K) or in peripheral blood (fig. S14L). Interestingly, a pro-angiogenic neutrophil type expressing genes linked to hypoxia and glycolysis in one of these studies (referred to as TAN-1 (55)) strongly matched the T3 state in the murine tumor, indicative that our T3 classification can capture subsets independently identified to be pro-tumoral (fig. S15A). Similarly, tumor neutrophil gene signatures derived from our mouse dataset (fig. S15B) were conserved in their ability to distinguish between human tumor neutrophils, and showed strong specificity in identifying T3, T2 and T1 neutrophils within the tumor and not in the adjacent normal tissue (fig. S15B). In all, our findings suggest that tumor-induced reprogramming of neutrophils is conserved in humans.
Because T3 neutrophils promote the growth of pancreatic tumors, we hypothesized that the genetic signature associated with the T3 state might be predictive of pancreatic cancer outcomes in human patients. To test this, we performed survival analysis on two independent pancreatic cancer cohorts from The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium Pancreatic Cancer-Australia (PACA-AU) by scoring patients based on high and low expression of each signature (see Methods for scoring criteria). Patients with high expression of the T3 neutrophil signature had poorer overall survival across both data sets, with a median of 652 (TCGA) and 427 (PACA-AU) days respectively (Fig. 6K), and this was independent of potential confounders such as patient gender, age and tumor stage (Table S5). Similarly, when disease-free survival (DFS, assessed as time to an adverse event from the initial treatment) was evaluated, patients with high T3 neutrophil signature expression had reduced DFS (Fig. 6M), with equal distribution of potential confounders across the two groups (Table S6). When T1 and T2 signatures were considered, high T2 signature expression correlated with worse OS only in the TCGA dataset (fig. S15C–D) but this was confounded by gender (Table S5), while high T1 signature expression was associated with lower DFS only in the PACA-AU dataset (fig. S15E–F). Hence, only the T3 signature was consistently associated with poorer OS and DFS across both datasets. We next considered whether the T3 neutrophil signature was also associated with poorer overall survival in other solid tumors. Within the TCGA pan-cancer database (56), higher expression of the T3 signature was associated with a significantly higher risk of death across a subset of solid cancers, including pancreatic cancer (PAAD), (Fig. 6L, Table S7 for p-values and confidence intervals). In contrast, T2 and T1 signatures were both protective against (lower hazard ratios, HRs) and associated with (higher HRs) patient death depending on the solid tumor (Fig. 6L, Table S7), in line with their nature as transitional subsets in the process of reprogramming. Taken together, these data support a model where tumor-educated T3 neutrophils drive tumor progression both in mouse and human cancers.
Discussion
Environmental cues can fine-tune immune responses by inducing cell recruitment and expansion of immune cells, generating productive responses with adequate numbers and specialized phenotypes. Unable to further proliferate, neutrophils rely on their ability to swiftly mobilize into tissues to perform their functions effectively and, in diseases like cancer, neutrophils at various maturation stages, tissue origins, and phenotypes are recruited into the tumor in large numbers (12–14, 17, 19). Given the likely co-existence of multiple tumor neutrophil states with different functional phenotypes, it is thus unclear how neutrophils exert a focused local effect in the tumor. Here, we examine how neutrophils decouple their initial maturation phenotype from their eventual pro-tumoral function by undergoing convergent reprograming within the tumor.
Our study demonstrates that tumor-infiltrating immature and mature neutrophils acquire distinct epigenetic, transcriptomic, and proteomic phenotypes while retaining features of their initial maturation status. We find that both immature (T1) and mature (T2) tumor neutrophils within the tumor converge towards a third population (T3) expressing dcTRAIl-R1. T3 neutrophils thus show intermediate maturation scores when quantified at the population level, and have both toroidal (immature) and hyper-segmented (mature) neutrophil nuclei, representative of them being an admixture of reprogrammed neutrophils of T1 and T2 origin. Our data reveals the capacity of neutrophils to simultaneously integrate different types of signal from the environment, allowing them to layer a new functional phenotype onto their pre-existing differentiation stage. We demonstrated this plasticity with immature and mature neutrophils from tumor-naïve mice, which are both capable of acquiring phenotypic (dcTRAIL-R1 expression) and transcriptional traits of the T3 state when cultured in tumor-conditioned media or upon entering the tumor in vivo. By bypassing neutrophil maturation as a rate-limiting step, this adaptability allows immature neutrophils to be equally mobilized and reprogrammed within the tumor within a shorter timeframe. Subsequently, this intrinsic ability embedded in neutrophils consolidates the various functional neutrophil states into one terminal neutrophil phenotype as directed by the tissue, in this case, a tumor.
Circulating neutrophils have a predicted half-life of ~10 hours (57), while tissue-resident neutrophils persist for up to one day (48). Although studies have hinted that neutrophils persist far longer within the tumor (10, 58, 59), whether this coincides with terminal differentiation in the tumor remains an open question. Using a BrdU pulse-labelling approach, we revealed that up to 5% of the originally labelled neutrophils could remain within the tumors for as long as 5.625 days upon entry. In addition, while our curve-fit was accurate for the first labelling timepoints, there was a noticeable underfitting at 12- and 15-days post-labelling, representing a possible underestimation of the full tumor neutrophil lifespan and indicating that a small population of neutrophils could persist even longer within the tumor. Long-lived neutrophils were predominantly of the dcTRAIL-R1hi T3 phenotype, which indicates a correlation between their ability to survive within challenging hypoxic and glycolytic environments and their continued persistence within the tumor. Spatial mapping at the transcriptome and protein level placed T3 neutrophils predominantly within a hypoxic-glycolytic niche nearer to the tumor core. In contrast, T1 and T2 neutrophils were positioned at the stromal and tumor parenchyma where a large vessel network exists, supporting the notion that these cell states are in the process of reprogramming after tumor entry. Given that hypoxia is not required to trigger T3 reprogramming, which can occur in normoxic conditions, we propose that migration towards the hypoxic-glycolytic niche occurs after acquisition of T3 epigenetic and transcriptional programs is fully complete to ensure neutrophil survival. Consistent with this possibility, upstream transcription factors regulating metabolic and oxidative stress are switched on in T3 neutrophils compared to T1 or T2 populations. Deletion or inhibiting these transcription factors to trace the timeline of T3 reprogramming thus represent important goals for future studies within the field.
T3 neutrophils accelerated early tumor growth experimentally, and while we still observed tumor growth with other introduced neutrophil subsets, this occurred at a slower rate likely due to recruitment and reprogramming of endogenous T3 neutrophils at the later timepoints. Notably, T1 and T2 neutrophils did not show statistically significant enhancement or inhibition of tumor growth compared to other control neutrophil populations or PBS, reflecting their transitional nature as opposed to a fully anti- or pro-tumoral population. In contrast, the sustained growth advantage conferred by T3 neutrophils stemmed, at least in part, from T3-dependent remodeling of the vasculature towards the core of the tumor. Given their localization within the glycolytic-hypoxic niche, an intriguing scenario would see T3 neutrophils serve as possible guide-rails to direct angiogenesis to relieve hypoxic and nutrient stress in areas that would most require it. In accordance, resistance towards anti-angiogenic therapies in human cancer has been associated with neutrophil infiltration (60), and neutrophil depletion reduces tumor vascularization and growth (50, 51, 61, 62). Studies facilitating further understanding of T3-mediated vascular remodelling would reveal new therapeutic targets against pathological angiogenesis within the tumor.
A particularly striking finding is the conservation of this differentiation program in tumor-infiltrating neutrophils across tumor type and species. When scRNAseq datasets from other tumor models were examined, T3 annotation could identify pro-tumoral clusters found both in mouse (i.e. mN5, (11)) and in human (i.e. TAN-1,(55)). T3 neutrophils promoted PDAC tumor growth in mice, while ablation of T3 neutrophils or their pro-angiogenic function removed this growth advantage. In parallel, the T3 signature consistently predicted poorer patient outcomes in two independent human PDAC cohorts, as well as in a subset of other solid tumors. Thus, different studies converge upon our identification of a terminally differentiated neutrophil state, whose signature can now be used to better understand neutrophil function in cancer and to predict tumor progression. We propose that the reported heterogeneity of neutrophils across tumors more likely reflects transitional states derived from populations at different stages of maturation and/or reprogramming.
Collectively, by ordering neutrophils through the lens of their ontogeny, we assess global neutrophil phenotypic heterogeneity through maturation stages, extra-medullary sources, and the specialization of each neutrophil to each tissue (48). Although all of these states co-exist within the tumor-bearing mouse, the neutrophil maturation trajectory remains unchanged, consistent with findings in other pro-inflammatory settings (19, 20, 63, 64). As such, while our findings do not fully exclude the possibility of neutrophil reprogramming outside of the tumor, they suggest that it is unlikely that upstream changes in neutrophil progenitors specifically drive the formation of a pro-tumoral neutrophil population. Instead, it is the intrinsic capacity of recruited neutrophils to adapt in response to the tumor environment, regardless of their initial phenotype, that allows them to adopt convergent trajectories to settle upon a final, pro-tumoral state. This feed-forward loop thus ensures the continued supply and differentiation of long-lived, pro-angiogenic neutrophils that support tumor growth. Expanding from the results of our study, we propose that it is advantageous for tissues to induce functional homogeneity in neutrophils at the local scale to support tissue growth and function. This process is then hijacked by the tumor to favor a functional neutrophil state that promotes aberrant tumor growth (fig. S16). Our findings thus reveal a general mechanism by which short-lived effector cells such as neutrophils efficiently adjust their functions to meet the demands of a tissue, and suggests that local neutrophil responses can be therapeutically targeted.
Supplementary Material
Acknowledgments:
We acknowledge and thank Matteo Iannacone and all members of the L.G.N. laboratory for helpful discussion and feedback on the manuscript. We thank the SIgN Flow Cytometry team for sorting and flow cytometry assistance, the SIgN Immunogenomics team for their assistance with generating and running the ATACseq and scRNAseq libraries, and the SIgN mouse core facility for their technical help and support.
Funding:
This research was funded by Singapore Immunology Network (SIgN) core funding and A*STAR, Singapore. N.L.G. is supported by core funding by SIgN. M.S.F is supported by the A*STAR Career Development Fund (202D8150). I.W.K. is supported by the A*STAR Career Development Fund (202D8197). S.Z.C. is supported by the A*STAR Career Development Award (192D8043) and core funding from SIgN. Y.T. and H.L.T. is supported by the Clinician Scientist Awards (NMRC/CSA-INV/0023/2017 and CSAINV20nov-0003) from the National Medical Research Council of Singapore. The SIgN Flow Cytometry facility is supported by National Research Foundation (NRF) Singapore under Shared Infrastructure Support (SIS) (NRF2017_SISFP09). For the use of the Ultramicroscope II, we would like to thank A*STAR core funds, the HMBS IAF-PP grant (H1701a0004) and the National Research Foundation’s Shared Infrastructure Support grant for SingaScope - a Singapore-wide microscopy infrastructure network (NRF2017_SISFP10) for continued support of the A*STAR Microscopy Platform. D.C-W. is supported by the CRI Irvington postdoctoral fellowship (CRI3511). A.H. was supported by grant R01AI165661 from NIH/NIAD, RTI2018-095497-B-I00 from MCIN, HR17_00527 from Fundación La Caixa, the Transatlantic Network of Excellence (TNE-18CVD04) from the Leducq Foundation, and FET-OPEN (no. 861878) from the European Commission. The CNIC is supported by the MCIN and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (CEX2020-001041-S).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability:
All data needed to evaluate the conclusions in the paper are present in the paper and/or in the Supplementary Materials. scRNAseq, ATACseq and spatial transcriptomics data in the paper will be eventually deposited in the NCBI Gene Expression Omnibus.
References
- 1.Ng LG, Ostuni R, Hidalgo A, Heterogeneity of neutrophils. Nat Rev Immunol. 19, 255–265 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, Diehn M, West RB, Plevritis SK, Alizadeh AA, The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 21, 938–945 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronte V, Brandau S, Chen S-H, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Rodriguez PC, Sica A, Umansky V, Vonderheide RH, Gabrilovich DI, Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun. 7, 12150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sagiv JY, Michaeli J, Assi S, Mishalian I, Kisos H, Levy L, Damti P, Lumbroso D, Polyansky L, Sionov RV, Ariel A, Hovav A-H, Henke E, Fridlender ZG, Granot Z, Phenotypic Diversity and Plasticity in Circulating Neutrophil Subpopulations in Cancer. Cell Reports. 10, 562–573 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Hsu BE, Tabariès S, Johnson RM, Andrzejewski S, Senecal J, Lehuédé C, Annis MG, Ma EH, Völs S, Ramsay L, Froment R, Monast A, Watson IR, Granot Z, Jones RG, St-Pierre J, Siegel PM, Immature Low-Density Neutrophils Exhibit Metabolic Flexibility that Facilitates Breast Cancer Liver Metastasis. Cell Reports. 27, 3902–3915.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Youn J-I, Nagaraj S, Collazo M, Gabrilovich DI, Subsets of Myeloid-Derived Suppressor Cells in Tumor-Bearing Mice. J Immunol. 181, 5791–5802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condamine T, Dominguez GA, Youn J-I, Kossenkov AV, Mony S, Alicea-Torres K, Tcyganov E, Hashimoto A, Nefedova Y, Lin C, Partlova S, Garfall A, Vogl DT, Xu X, Knight SC, Malietzis G, Lee GH, Eruslanov E, Albelda SM, Wang X, Mehta JL, Bewtra M, Rustgi A, Hockstein N, Witt R, Masters G, Nam B, Smirnov D, Sepulveda MA, Gabrilovich DI, Lectin-type oxidized LDL receptor-1 distinguishes population of human polymorphonuclear myeloid-derived suppressor cells in cancer patients. Sci. Immunol. 1 (2016), doi: 10.1126/sciimmunol.aaf8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engblom C, Pfirschke C, Zilionis R, Da Silva Martins J, Bos SA, Courties G, Rickelt S, Severe N, Baryawno N, Faget J, Savova V, Zemmour D, Kline J, Siwicki M, Garris C, Pucci F, Liao H-W, Lin Y-J, Newton A, Yaghi OK, Iwamoto Y, Tricot B, Wojtkiewicz GR, Nahrendorf M, Cortez-Retamozo V, Meylan E, Hynes RO, Demay M, Klein A, Bredella MA, Scadden DT, Weissleder R, Pittet MJ, Osteoblasts remotely supply lung tumors with cancer-promoting SiglecF high neutrophils. Science. 358, eaal5081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veglia F, Tyurin VA, Blasi M, De Leo A, Kossenkov AV, Donthireddy L, To TKJ, Schug Z, Basu S, Wang F, Ricciotti E, DiRusso C, Murphy ME, Vonderheide RH, Lieberman PM, Mulligan C, Nam B, Hockstein N, Masters G, Guarino M, Lin C, Nefedova Y, Black P, Kagan VE, Gabrilovich DI, Fatty acid transport protein 2 reprograms neutrophils in cancer. Nature. 569, 73–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfirschke C, Engblom C, Gungabeesoon J, Lin Y, Rickelt S, Zilionis R, Messemaker M, Siwicki M, Gerhard GM, Kohl A, Meylan E, Weissleder R, Klein AM, Pittet MJ, Tumor-Promoting Ly-6G+ SiglecFhigh Cells Are Mature and Long-Lived Neutrophils. Cell Reports. 32, 108164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV, Kerwin CM, Choi S, Richards WG, De Rienzo A, Tenen DG, Bueno R, Levantini E, Pittet MJ, Klein AM, Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity. 50, 1317–1334.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alshetaiwi H, Pervolarakis N, McIntyre LL, Ma D, Nguyen Q, Rath JA, Nee K, Hernandez G, Evans K, Torosian L, Silva A, Walsh C, Kessenbrock K, Defining the emergence of myeloid-derived suppressor cells in breast cancer using single-cell transcriptomics. Sci. Immunol. 5, eaay6017 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veglia F, Hashimoto A, Dweep H, Sanseviero E, De Leo A, Tcyganov E, Kossenkov A, Mulligan C, Nam B, Masters G, Patel J, Bhargava V, Wilkinson P, Smirnov D, Sepulveda MA, Singhal S, Eruslanov EB, Cristescu R, Loboda A, Nefedova Y, Gabrilovich DI, Analysis of classical neutrophils and polymorphonuclear myeloid-derived suppressor cells in cancer patients and tumor-bearing mice. Journal of Experimental Medicine. 218, e20201803 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evrard M, Kwok IWH, Chong SZ, Teng KWW, Becht E, Chen J, Sieow JL, Penny HL, Ching GC, Devi S, Adrover JM, Li JLY, Liong KH, Tan L, Poon Z, Foo S, Chua JW, Su I-H, Balabanian K, Bachelerie F, Biswas SK, Larbi A, Hwang WYK, Madan V, Koeffler HP, Wong SC, Newell EW, Hidalgo A, Ginhoux F, Ng LG, Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity. 48, 364–379.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Zhu YP, Padgett L, Dinh HQ, Marcovecchio P, Blatchley A, Wu R, Ehinger E, Kim C, Mikulski Z, Seumois G, Madrigal A, Vijayanand P, Hedrick CC, Identification of an Early Unipotent Neutrophil Progenitor with Pro-tumoral Activity in Mouse and Human Bone Marrow. Cell Reports. 24, 2329–2341.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwok I, Becht E, Xia Y, Ng M, Teh YC, Tan L, Evrard M, Li JLY, Tran HTN, Tan Y, Liu D, Mishra A, Liong KH, Leong K, Zhang Y, Olsson A, Mantri CK, Shyamsunder P, Liu Z, Piot C, Dutertre C-A, Cheng H, Bari S, Ang N, Biswas SK, Koeffler HP, Tey HL, Larbi A, Su I-H, Lee B, John A. St., Chan JKY, Hwang WYK, Chen J, Salomonis N, Chong SZ, Grimes HL, Liu B, Hidalgo A, Newell EW, Cheng T, Ginhoux F, Ng LG, Combinatorial Single-Cell Analyses of Granulocyte-Monocyte Progenitor Heterogeneity Reveals an Early Uni-potent Neutrophil Progenitor. Immunity. 53, 303–318.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Dinh HQ, Eggert T, Meyer MA, Zhu YP, Olingy CE, Llewellyn R, Wu R, Hedrick CC, Coexpression of CD71 and CD117 Identifies an Early Unipotent Neutrophil Progenitor Population in Human Bone Marrow. Immunity. 53, 319–334.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muench DE, Olsson A, Ferchen K, Pham G, Serafin RA, Chutipongtanate S, Dwivedi P, Song B, Hay S, Chetal K, Trump-Durbin LR, Mookerjee-Basu J, Zhang K, Yu JC, Lutzko C, Myers KC, Nazor KL, Greis KD, Kappes DJ, Way SS, Salomonis N, Grimes HL, Mouse models of neutropenia reveal progenitor-stage-specific defects. Nature. 582, 109–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie X, Shi Q, Wu P, Zhang X, Kambara H, Su J, Yu H, Park S-Y, Guo R, Ren Q, Zhang S, Xu Y, Silberstein LE, Cheng T, Ma F, Li C, Luo HR, Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat Immunol. 21, 1119–1133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoyratty TE, Ai Z, Ballesteros I, Eames HL, Mathie S, Martín-Salamanca S, Wang L, Hemmings A, Willemsen N, von Werz V, Zehrer A, Walzog B, van Grinsven E, Hidalgo A, Udalova IA, Distinct transcription factor networks control neutrophil-driven inflammation. Nat Immunol. 22, 1093–1106 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu C, Ning H, Liu M, Lin J, Luo S, Zhu W, Xu J, Wu W-C, Liang J, Shao C-K, Ren J, Wei B, Cui J, Chen M-S, Zheng L, Spleen mediates a distinct hematopoietic progenitor response supporting tumor-promoting myelopoiesis. Journal of Clinical Investigation. 128, 3425–3438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu M, Wu C, Luo S, Hua Q, Chen H-T, Weng Y, Xu J, Lin H, Wang L, Li J, Zhu L, Guo Z, Zhuang S-M, Kang T, Zheng L, PERK reprograms hematopoietic progenitor cells to direct tumor-promoting myelopoiesis in the spleen. Journal of Experimental Medicine. 219, e20211498 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S, Tuveson DA, Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 7, 469–483 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Angerer P, Haghverdi L, Büttner M, Theis FJ, Marr C, Buettner F, destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics. 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ, Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 13, 845–848 (2016). [DOI] [PubMed] [Google Scholar]
- 26.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, Fan J, Borm LE, Liu Z, van Bruggen D, Guo J, He X, Barker R, Sundström E, Castelo-Branco G, Cramer P, Adameyko I, Linnarsson S, Kharchenko PV, RNA velocity of single cells. Nature. 560, 494–498 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van de Sande B, Flerin C, Davie K, De Waegeneer M, Hulselmans G, Aibar S, Seurinck R, Saelens W, Cannoodt R, Rouchon Q, Verbeiren T, De Maeyer D, Reumers J, Saeys Y, Aerts S, A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 15, 2247–2276 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Dutertre C-A, Becht E, Irac SE, Khalilnezhad A, Narang V, Khalilnezhad S, Ng PY, van den Hoogen LL, Leong JY, Lee B, Chevrier M, Zhang XM, Yong PJA, Koh G, Lum J, Howland SW, Mok E, Chen J, Larbi A, Tan HKK, Lim TKH, Karagianni P, Tzioufas AG, Malleret B, Brody J, Albani S, van Roon J, Radstake T, Newell EW, Ginhoux F, Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity. 51, 573–589.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Becht E, Tolstrup D, Dutertre C-A, Morawski PA, Campbell DJ, Ginhoux F, Newell EW, Gottardo R, Headley MB, High-throughput single-cell quantification of hundreds of proteins using conventional flow cytometry and machine learning. Sci. Adv. 7, eabg0505 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neumann K, Castiñeiras-Vilariño M, Höckendorf U, Hannesschläger N, Lemeer S, Kupka D, Meyermann S, Lech M, Anders H-J, Kuster B, Busch DH, Gewies A, Naumann R, Groß O, Ruland J, Clec12a Is an Inhibitory Receptor for Uric Acid Crystals that Regulates Inflammation in Response to Cell Death. Immunity. 40, 389–399 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Xu W, Dong J, Zheng Y, Zhou J, Yuan Y, Ta HM, Miller HE, Olson M, Rajasekaran K, Ernstoff MS, Wang D, Malarkannan S, Wang L, Immune-Checkpoint Protein VISTA Regulates Antitumor Immunity by Controlling Myeloid Cell–Mediated Inflammation and Immunosuppression. Cancer Immunology Research. 7, 1497–1510 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryzhov SV, Pickup MW, Chytil A, Gorska AE, Zhang Q, Owens P, Feoktistov I, Moses HL, Novitskiy SV, Role of TGF-β Signaling in Generation of CD39 + CD73 + Myeloid Cells in Tumors. J.I. 193, 3155–3164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, Brown M, Chang C, Troncoso L, Chen ST, Sastre-Perona A, Park MD, Tabachnikova A, Dhainaut M, Hamon P, Maier B, Sawai CM, Agulló-Pascual E, Schober M, Brown BD, Reizis B, Marron T, Kenigsberg E, Moussion C, Benaroch P, Aguirre-Ghiso JA, Merad M, Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature. 595, 578–584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaur S, Bronson SM, Pal-Nath D, Miller TW, Soto-Pantoja DR, Roberts DD, Functions of Thrombospondin-1 in the Tumor Microenvironment. IJMS. 22, 4570 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Markowska AI, Liu F-T, Panjwani N, Galectin-3 is an important mediator of VEGF- and bFGF-mediated angiogenic response. Journal of Experimental Medicine. 207, 1981–1993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao E, Stone MR, Ren X, Guenthoer J, Smythe KS, Pulliam T, Williams SR, Uytingco CR, Taylor SEB, Nghiem P, Bielas JH, Gottardo R, Spatial transcriptomics at subspot resolution with BayesSpace. Nat Biotechnol. 39, 1375–1384 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu W, Kang Y, Epithelial-Mesenchymal Plasticity in Cancer Progression and Metastasis. Developmental Cell. 49, 361–374 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleshchevnikov V, Shmatko A, Dann E, Aivazidis A, King HW, Li T, Elmentaite R, Lomakin A, Kedlian V, Gayoso A, Jain MS, Park JS, Ramona L, Tuck E, Arutyunyan A, Vento-Tormo R, Gerstung M, James L, Stegle O, Bayraktar OA, Cell2location maps fine-grained cell types in spatial transcriptomics. Nat Biotechnol. 40, 661–671 (2022). [DOI] [PubMed] [Google Scholar]
- 39.Kinkhabwala A, Herbel C, Pankratz J, Yushchenko DA, Rüberg S, Praveen P, Reiß S, Rodriguez FC, Schäfer D, Kollet J, Dittmer V, Martinez-Osuna M, Minnerup L, Reinhard C, Dzionek A, Rockel TD, Borbe S, Büscher M, Krieg J, Nederlof M, Jungblut M, Eckardt D, Hardt O, Dose C, Schumann E, Peters R-P, Miltenyi S, Schmitz J, Müller W, Bosio A, MACSima imaging cyclic staining (MICS) technology reveals combinatorial target pairs for CAR T cell treatment of solid tumors. Sci Rep. 12, 1911 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kinugasa Y, Matsui T, Takakura N, CD44 Expressed on Cancer-Associated Fibroblasts Is a Functional Molecule Supporting the Stemness and Drug Resistance of Malignant Cancer Cells in the Tumor Microenvironment. Stem Cells. 32, 145–156 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Costa A, Kieffer Y, Scholer-Dahirel A, Pelon F, Bourachot B, Cardon M, Sirven P, Magagna I, Fuhrmann L, Bernard C, Bonneau C, Kondratova M, Kuperstein I, Zinovyev A, Givel A-M, Parrini M-C, Soumelis V, Vincent-Salomon A, Mechta-Grigoriou F, Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell. 33, 463–479.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Slack RJ, Mills R, Mackinnon AC, The therapeutic potential of galectin-3 inhibition in fibrotic disease. The International Journal of Biochemistry & Cell Biology. 130, 105881 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Fonsatti E, Altomonte M, Nicotra MR, Natali PG, Maio M, Endoglin (CD105): a powerful therapeutic target on tumor-associated angiogenetic blood vessels. Oncogene. 22, 6557–6563 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Giatromanolaki A, Kouroupi M, Pouliliou S, Mitrakas A, Hasan F, Pappa A, Koukourakis MI, Ectonucleotidase CD73 and CD39 expression in non-small cell lung cancer relates to hypoxia and immunosuppressive pathways. Life Sciences. 259, 118389 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Losenkova K, Zuccarini M, Karikoski M, Laurila J, Boison D, Jalkanen S, Yegutkin GG, Journal of Cell Science, in press, doi: 10.1242/jcs.241463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petruk N, Tuominen S, Åkerfelt M, Mattsson J, Sandholm J, Nees M, Yegutkin GG, Jukkola A, Tuomela J, Selander KS, CD73 facilitates EMT progression and promotes lung metastases in triple-negative breast cancer. Sci Rep. 11, 6035 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Z, Chen S, Zhu Y, Zhang D, Xie P, Jiao Q, Chi J, Xu S, Xue Y, Lu X, Song X, Cristofanilli M, Gradishar WJ, Kalinsky K, Yin Y, Zhang B, Wan Y, Proteolytic regulation of CD73 by TRIM21 orchestrates tumor immunogenicity. Sci. Adv. 9, eadd6626 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballesteros I, Rubio-Ponce A, Genua M, Lusito E, Kwok I, Fernández-Calvo G, Khoyratty TE, van Grinsven E, González-Hernández S, Nicolás-Ávila JÁ, Vicanolo T, Maccataio A, Benguría A, Li JL, Adrover JM, Aroca-Crevillen A, Quintana JA, Martín-Salamanca S, Mayo F, Ascher S, Barbiera G, Soehnlein O, Gunzer M, Ginhoux F, Sánchez-Cabo F, Nistal-Villán E, Schulz C, Dopazo A, Reinhardt C, Udalova IA, Ng LG, Ostuni R, Hidalgo A, Co-option of Neutrophil Fates by Tissue Environments. Cell. 183, 1282–1297.e18 (2020). [DOI] [PubMed] [Google Scholar]
- 49.Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, Matrisian LM, Carbone DP, Lin PC, Expansion of myeloid immune suppressor GrϩCD11bϩ cells in tumor-bearing host directly promotes tumor angiogenesis. CANCER CELL, 13 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Nozawa H, Chiu C, Hanahan D, Infiltrating neutrophils mediate the initial angiogenic switch in a mouse model of multistage carcinogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 12493–12498 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shojaei F, Wu X, Zhong C, Yu L, Liang X-H, Yao J, Blanchard D, Bais C, Peale FV, van Bruggen N, Ho C, Ross J, Tan M, Carano RAD, Meng YG, Ferrara N, Bv8 regulates myeloid-cell-dependent tumour angiogenesis. Nature. 450, 825–831 (2007). [DOI] [PubMed] [Google Scholar]
- 52.Kastana P, Zahra FT, Ntenekou D, Katraki-Pavlou S, Beis D, Lionakis MS, Mikelis CM, Papadimitriou E, “Matrigel Plug Assay for In Vivo Evaluation of Angiogenesis” in The Extracellular Matrix, Vigetti D, Theocharis AD, Eds. (Springer New York, New York, NY, 2019; http://link.springer.com/10.1007/978-1-4939-9133-4_18), vol. 1952 of Methods in Molecular Biology, pp. 219–232. [DOI] [PubMed] [Google Scholar]
- 53.Baeriswyl V, Christofori G, The angiogenic switch in carcinogenesis. Seminars in Cancer Biology. 19, 329–337 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L, Lazarus J, Amir ED, Gunchick V, Espinoza C, Bell S, Harris L, Lima F, Irizarry-Negron V, Paglia D, Macchia J, Chu AKY, Schofield H, Wamsteker E-J, Kwon R, Schulman A, Prabhu A, Law R, Sondhi A, Yu J, Patel A, Donahue K, Nathan H, Cho C, Anderson MA, Sahai V, Lyssiotis CA, Zou W, Allen BL, Rao A, Crawford HC, Bednar F, Frankel TL, Pasca di Magliano M, Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat Cancer. 1, 1097–1112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Liu Y, Dai Y, Tang X, Yin T, Wang C, Wang T, Dong L, Shi M, Qin J, Xue M, Cao Y, Liu J, Liu P, Huang J, Wen C, Zhang J, Xu Z, Bai F, Deng X, Peng C, Chen H, Jiang L, Chen S, Shen B, Single-cell RNA-seq analysis reveals BHLHE40-driven pro-tumour neutrophils with hyperactivated glycolysis in pancreatic tumour microenvironment. Gut (2022), doi: 10.1136/gutjnl-2021-326070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H, Caesar-Johnson SJ, Demchok JA, Felau I, Kasapi M, Ferguson ML, Hutter CM, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Zhang J.(Julia), Chudamani S, Liu J, Lolla L, Naresh R, Pihl T, Sun Q, Wan Y, Wu Y, Cho J, DeFreitas T, Frazer S, Gehlenborg N, Getz G, Heiman DI, Kim J, Lawrence MS, Lin P, Meier S, Noble MS, Saksena G, Voet D, Zhang H, Bernard B, Chambwe N, Dhankani V, Knijnenburg T, Kramer R, Leinonen K, Liu Y, Miller M, Reynolds S, Shmulevich I, Thorsson V, Zhang W, Akbani R, Broom BM, Hegde AM, Ju Z, Kanchi RS, Korkut A, Li J, Liang H, Ling S, Liu W, Lu Y, Mills GB, Ng K-S, Rao A, Ryan M, Wang J, Weinstein JN, Zhang J, Abeshouse A, Armenia J, Chakravarty D, Chatila WK, De Bruijn I, Gao J, Gross BE, Heins ZJ, Kundra R, La K, Ladanyi M, Luna A, Nissan MG, Ochoa A, Phillips SM, Reznik E, Sanchez-Vega F, Sander C, Schultz N, Sheridan R, Sumer SO, Sun Y, Taylor BS, Wang J, Zhang H, Anur P, Peto M, Spellman P, Benz C, Stuart JM, Wong CK, Yau C, Hayes DN, Parker JS, Wilkerson MD, Ally A, Balasundaram M, Bowlby R, Brooks D, Carlsen R, Chuah E, Dhalla N, Holt R, Jones SJM, Kasaian K, Lee D, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall K, Robertson AG, Sadeghi S, Schein JE, Sipahimalani P, Tam A, Thiessen N, Tse K, Wong T, Berger AC, Beroukhim R, Cherniack AD, Cibulskis C, Gabriel SB, Gao GF, Ha G, Meyerson M, Schumacher SE, Shih J, Kucherlapati MH, Kucherlapati RS, Baylin S, Cope L, Danilova L, Bootwalla MS, Lai PH, Maglinte DT, Van Den Berg DJ, Weisenberger DJ, Auman JT, Balu S, Bodenheimer T, Fan C, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Meng S, Mieczkowski PA, Mose LE, Perou AH, Perou CM, Roach J, Shi Y, Simons JV, Skelly T, Soloway MG, Tan D, Veluvolu U, Fan H, Hinoue T, Laird PW, Shen H, Zhou W, Bellair M, Chang K, Covington K, Creighton CJ, Dinh H, Doddapaneni H, Donehower LA, Drummond J, Gibbs RA, Glenn R, Hale W, Han Y, Hu J, Korchina V, Lee S, Lewis L, Li W, Liu X, Morgan M, Morton D, Muzny D, Santibanez J, Sheth M, Shinbro E, Wang L, Wang M, Wheeler DA, Xi L, Zhao F, Hess J, Appelbaum EL, Bailey M, Cordes MG, Ding L, Fronick CC, Fulton LA, Fulton RS, Kandoth C, Mardis ER, McLellan MD, Miller CA, Schmidt HK, Wilson RK, Crain D, Curley E, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Thompson E, Yena P, Bowen J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Corcoran N, Costello T, Hovens C, Carvalho AL, De Carvalho AC, Fregnani JH, Longatto-Filho A, Reis RM, Scapulatempo-Neto C, Silveira HCS, Vidal DO, Burnette A, Eschbacher J, Hermes B, Noss A, Singh R, Anderson ML, Castro PD, Ittmann M, Huntsman D, Kohl B, Le X, Thorp R, Andry C, Duffy ER, Lyadov V, Paklina O, Setdikova G, Shabunin A, Tavobilov M, McPherson C, Warnick R, Berkowitz R, Cramer D, Feltmate C, Horowitz N, Kibel A, Muto M, Raut CP, Malykh A, Barnholtz-Sloan JS, Barrett W, Devine K, Fulop J, Ostrom QT, Shimmel K, Wolinsky Y, Sloan AE, De Rose A, Giuliante F, Goodman M, Karlan BY, Hagedorn CH, Eckman J, Harr J, Myers J, Tucker K, Zach LA, Deyarmin B, Hu H, Kvecher L, Larson C, Mural RJ, Somiari S, Vicha A, Zelinka T, Bennett J, Iacocca M, Rabeno B, Swanson P, Latour M, Lacombe L, Têtu B, Bergeron A, McGraw M, Staugaitis SM, Chabot J, Hibshoosh H, Sepulveda A, Su T, Wang T, Potapova O, Voronina O, Desjardins L, Mariani O, Roman-Roman S, Sastre X, Stern M-H, Cheng F, Signoretti S, Berchuck A, Bigner D, Lipp E, Marks J, McCall S, McLendon R, Secord A, Sharp A, Behera M, Brat DJ, Chen A, Delman K, Force S, Khuri F, Magliocca K, Maithel S, Olson JJ, Owonikoko T, Pickens A, Ramalingam S, Shin DM, Sica G, Van Meir EG, Zhang H, Eijckenboom W, Gillis A, Korpershoek E, Looijenga L, Oosterhuis W, Stoop H, Van Kessel KE, Zwarthoff EC, Calatozzolo C, Cuppini L, Cuzzubbo S, DiMeco F, Finocchiaro G, Mattei L, Perin A, Pollo B, Chen C, Houck J, Lohavanichbutr P, Hartmann A, Stoehr C, Stoehr R, Taubert H, Wach S, Wullich B, Kycler W, Murawa D, Wiznerowicz M, Chung K, Edenfield WJ, Martin J, Baudin E, Bubley G, Bueno R, De Rienzo A, Richards WG, Kalkanis S, Mikkelsen T, Noushmehr H, Scarpace L, Girard N, Aymerich M, Campo E, Giné E, Guillermo AL, Van Bang N, Hanh PT, Phu BD, Tang Y, Colman H, Evason K, Dottino PR, Martignetti JA, Gabra H, Juhl H, Akeredolu T, Stepa S, Hoon D, Ahn K, Kang KJ, Beuschlein F, Breggia A, Birrer M, Bell D, Borad M, Bryce AH, Castle E, Chandan V, Cheville J, Copland JA, Farnell M, Flotte T, Giama N, Ho T, Kendrick M, Kocher J-P, Kopp K, Moser C, Nagorney D, O’Brien D, O’Neill BP, Patel T, Petersen G, Que F, Rivera M, Roberts L, Smallridge R, Smyrk T, Stanton M, Thompson RH, Torbenson M, Yang JD, Zhang L, Brimo F, Ajani JA, Angulo Gonzalez AM, Behrens C, Bondaruk J, Broaddus R, Czerniak B, Esmaeli B, Fujimoto J, Gershenwald J, Guo C, Lazar AJ, Logothetis C, Meric-Bernstam F, Moran C, Ramondetta L, Rice D, Sood A, Tamboli P, Thompson T, Troncoso P, Tsao A, Wistuba I, Carter C, Haydu L, Hersey P, Jakrot V, Kakavand H, Kefford R, Lee K, Long G, Mann G, Quinn M, Saw R, Scolyer R, Shannon K, Spillane A, Stretch J, Synott M, Thompson J, Wilmott J, Al-Ahmadie H, Chan TA, Ghossein R, Gopalan A, Levine DA, Reuter V, Singer S, Singh B, Tien NV, Broudy T, Mirsaidi C, Nair P, Drwiega P, Miller J, Smith J, Zaren H, Park J-W, Hung NP, Kebebew E, Linehan WM, Metwalli AR, Pacak K, Pinto PA, Schiffman M, Schmidt LS, Vocke CD, Wentzensen N, Worrell R, Yang H, Moncrieff M, Goparaju C, Melamed J, Pass H, Botnariuc N, Caraman I, Cernat M, Chemencedji I, Clipca A, Doruc S, Gorincioi G, Mura S, Pirtac M, Stancul I, Tcaciuc D, Albert M, Alexopoulou I, Arnaout A, Bartlett J, Engel J, Gilbert S, Parfitt J, Sekhon H, Thomas G, Rassl DM, Rintoul RC, Bifulco C, Tamakawa R, Urba W, Hayward N, Timmers H, Antenucci A, Facciolo F, Grazi G, Marino M, Merola R, De Krijger R, Gimenez-Roqueplo A-P, Piché A, Chevalier S, McKercher G, Birsoy K, Barnett G, Brewer C, Farver C, Naska T, Pennell NA, Raymond D, Schilero C, Smolenski K, Williams F, Morrison C, Borgia JA, Liptay MJ, Pool M, Seder CW, Junker K, Omberg L, Dinkin M, Manikhas G, Alvaro D, Bragazzi MC, Cardinale V, Carpino G, Gaudio E, Chesla D, Cottingham S, Dubina M, Moiseenko F, Dhanasekaran R, Becker K-F, Janssen K-P, Slotta-Huspenina J, Abdel-Rahman MH, Aziz D, Bell S, Cebulla CM, Davis A, Duell R, Elder JB, Hilty J, Kumar B, Lang J, Lehman NL, Mandt R, Nguyen P, Pilarski R, Rai K, Schoenfield L, Senecal K, Wakely P, Hansen P, Lechan R, Powers J, Tischler A, Grizzle WE, Sexton KC, Kastl A, Henderson J, Porten S, Waldmann J, Fassnacht M, Asa SL, Schadendorf D, Couce M, Graefen M, Huland H, Sauter G, Schlomm T, Simon R, Tennstedt P, Olabode O, Nelson M, Bathe O, Carroll PR, Chan JM, Disaia P, Glenn P, Kelley RK, Landen CN, Phillips J, Prados M, Simko J, Smith-McCune K, VandenBerg S, Roggin K, Fehrenbach A, Kendler A, Sifri S, Steele R, Jimeno A, Carey F, Forgie I, Mannelli M, Carney M, Hernandez B, Campos B, Herold-Mende C, Jungk C, Unterberg A, Von Deimling A, Bossler A, Galbraith J, Jacobus L, Knudson M, Knutson T, Ma D, Milhem M, Sigmund R, Godwin AK, Madan R, Rosenthal HG, Adebamowo C, Adebamowo SN, Boussioutas A, Beer D, Giordano T, Mes-Masson A-M, Saad F, Bocklage T, Landrum L, Mannel R, Moore K, Moxley K, Postier R, Walker J, Zuna R, Feldman M, Valdivieso F, Dhir R, Luketich J, Mora Pinero EM, Quintero-Aguilo M, Carlotti CG Jr, Dos Santos JS, Kemp R, Sankarankuty A, Tirapelli D, Catto J, Agnew K, Swisher E, Creaney J, Robinson B, Shelley CS, Godwin EM, Kendall S, Shipman C, Bradford C, Carey T, Haddad A, Moyer J, Peterson L, Prince M, Rozek L, Wolf G, Bowman R, Fong KM, Yang I, Korst R, Rathmell WK, Fantacone-Campbell JL, Hooke JA, Kovatich AJ, Shriver CD, DiPersio J, Drake B, Govindan R, Heath S, Ley T, Van Tine B, Westervelt P, Rubin MA, Lee JI, Aredes ND, Mariamidze A, An Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 173, 400–416.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pillay J, den Braber I, Vrisekoop N, Kwast LM, de Boer RJ, Borghans JAM, Tesselaar K, Koenderman L, In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood. 116, 625–627 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Sawanobori Y, Ueha S, Kurachi M, Shimaoka T, Talmadge JE, Abe J, Shono Y, Kitabatake M, Kakimi K, Mukaida N, Matsushima K, Chemokine-mediated rapid turnover of myeloid-derived suppressor cells in tumor-bearing mice. Blood. 111, 5457–5466 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Ancey P-B, Contat C, Boivin G, Sabatino S, Pascual J, Zangger N, Perentes JY, Peters S, Abel ED, Kirsch DG, Rathmell JC, Vozenin M-C, Meylan E, GLUT1 Expression in Tumor-Associated Neutrophils Promotes Lung Cancer Growth and Resistance to Radiotherapy. Cancer Research. 81, 2345–2357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schiffmann LM, Fritsch M, Gebauer F, Günther SD, Stair NR, Seeger JM, Thangarajah F, Dieplinger G, Bludau M, Alakus H, Göbel H, Quaas A, Zander T, Hilberg F, Bruns CJ, Kashkar H, Coutelle O, Tumour-infiltrating neutrophils counteract anti-VEGF therapy in metastatic colorectal cancer. Br J Cancer. 120, 69–78 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jablonska J, Leschner S, Westphal K, Lienenklaus S, Weiss S, Neutrophils responsive to endogenous IFN-β regulate tumor angiogenesis and growth in a mouse tumor model. J. Clin. Invest. 120, 1151–1164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Itatani Y, Yamamoto T, Zhong C, Molinolo AA, Ruppel J, Hegde P, Taketo MM, Ferrara N, Suppressing neutrophil-dependent angiogenesis abrogates resistance to anti-VEGF antibody in a genetic model of colorectal cancer. Proc. Natl. Acad. Sci. U.S.A. 117, 21598–21608 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grieshaber-Bouyer R, Radtke FA, Cunin P, Stifano G, Levescot A, Vijaykumar B, Nelson-Maney N, Blaustein RB, Monach PA, Nigrovic PA, ImmGen Consortium, The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat Commun. 12, 2856 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montaldo E, Lusito E, Bianchessi V, Caronni N, Scala S, Basso-Ricci L, Cantaffa C, Masserdotti A, Barilaro M, Barresi S, Genua M, Vittoria FM, Barbiera G, Lazarevic D, Messina C, Xue E, Marktel S, Tresoldi C, Milani R, Ronchi P, Gattillo S, Santoleri L, Di Micco R, Ditadi A, Belfiori G, Aleotti F, Naldini MM, Gentner B, Gardiman E, Tamassia N, Cassatella MA, Hidalgo A, Kwok I, Ng LG, Crippa S, Falconi M, Pettinella F, Scapini P, Naldini L, Ciceri F, Aiuti A, Ostuni R, Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat Immunol. 23, 1470–1483 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or in the Supplementary Materials. scRNAseq, ATACseq and spatial transcriptomics data in the paper will be eventually deposited in the NCBI Gene Expression Omnibus.