Abstract
Gastric cancer (GC) is one of the most common gastrointestinal tumors. As a newly discovered type of non-coding RNAs, transfer RNA (tRNA)-derived small RNAs (tsRNAs) play a dual biological role in cancer. Our previous studies have demonstrated the potential of tRF-23-Q99P9P9NDD as a diagnostic and prognostic biomarker for GC. In this work, we confirmed for the first time that tRF-23-Q99P9P9NDD can promote the proliferation, migration, and invasion of GC cells in vitro. The dual luciferase reporter gene assay confirmed that tRF-23-Q99P9P9NDD could bind to the 3' untranslated region (UTR) site of acyl-coenzyme A dehydrogenase short/branched chain (ACADSB). In addition, ACADSB could rescue the effect of tRF-23-Q99P9P9NDD on GC cells. Next, we used Gene Ontology (GO), the Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) to find that downregulated ACADSB in GC may promote lipid accumulation by inhibiting fatty acid catabolism and ferroptosis. Finally, we verified the correlation between ACADSB and 12 ferroptosis genes at the transcriptional level, as well as the changes in reactive oxygen species (ROS) levels by flow cytometry. In summary, this study proposes that tRF-23-Q99P9P9NDD may affect GC lipid metabolism and ferroptosis by targeting ACADSB, thereby promoting GC progression. It provides a theoretical basis for the diagnostic and prognostic monitoring value of GC and opens up new possibilities for treatment.
Keywords: Transfer RNA (tRNA)-derived small RNA (tsRNA), Gastric cancer (GC), Acyl-coenzyme A dehydrogenase short/branched chain (ACADSB), Molecular mechanism, Treatment, Ferroptosis
Abstract
胃癌(GC)是最常见的胃肠道肿瘤之一。作为一种新型的非编码RNA,转移RNA(tRNA)衍生的小RNA(tsRNA)在肿瘤中发挥着双重生物学作用。我们之前的研究揭示了tRF-23-Q99P9P9NDD作为GC诊断和预后生物标志物的潜力。在本研究中,我们首次证实了tRF-23-Q99P9P9NDD能够促进GC细胞的增殖、迁移和侵袭。双荧光素酶报告基因实验证实tRF-23-Q99P9P9NDD可以结合短/支链酰基辅酶A脱氢酶(ACADSB)的3'非编码区(UTR)位点。此外,ACADSB可以挽救tRF-23-Q99P9P9NDD对GC细胞的影响。随后,我们使用基因本体论(GO)、京都基因和基因组百科全书(KEGG)以及基因集富集分析(GSEA)发现,GC中下调的ACADSB可能通过抑制脂肪酸分解代谢和铁死亡来促进脂质积累。最后,我们在转录水平上验证了ACADSB和12个铁死亡基因之间的相关性,并通过流式细胞仪检测了活性氧(ROS)水平的变化。综上,本研究提出tRF-23-Q99P9P9NDD可能通过靶向ACADSB影响GC脂质代谢和铁死亡,从而促进GC进展。这为GC的诊断和预后监测价值提供了理论基础,并为治疗开辟了新的可能性。
Keywords: tRNA衍生的小RNA(tsRNA), 胃癌(GC), 短/支链酰基辅酶A脱氢酶(ACADSB), 分子机制, 治疗, 铁死亡
1. Introduction
Gastric cancer (GC) is one of the most common cancers in the world. The incidence of GC has decreased with the advancement of medical treatments and the eradication of Helicobacter pylori. However, the mortality rate of GC still ranks fourth among cancers (Correa, 2013; Sung et al., 2021). The prognosis of GC remains poor as most patients are already in an advanced stage when diagnosed, often accompanied by lymph nodes or distant metastases (Shen et al., 2013). Consequently, it is urgent to identify biomarkers that can aid in the diagnosis and prognostic monitoring of GC and provide new treatment options for this disease.
With the development of next-generation sequencing technology, an emerging class of non-coding RNAs (ncRNAs), transfer RNA (tRNA)-derived small RNAs (tsRNAs), has been subject to extensive research. According to the cleavage sites of nucleases, tsRNAs are mainly divided into two categories: tRNA halves (tiRNAs) and tRNA-derived fragments (tRFs) (Liu et al., 2021; Xu et al., 2022). tiRNAs include 5' tiRNAs of approximately 31‒36 nucleotides (nt) in length and 3' tiRNAs of 36‒41 nt, which are usually generated under stress by angiopoietin (ANG) cleavage of the middle of the anticodon loop of mature tRNAs (Tao et al., 2020; Wen et al., 2021). tRFs are about 14‒30 nt similar to microRNAs (miRNAs), mainly generated through Dicer-dependent and -independent pathways. Depending on the enzymatic cleavage site, tRFs can be divided into tRF-1, tRF-3, tRF-5, and i-tRF (Kumar et al., 2016; Park and Kim, 2018; Yang et al., 2023). tsRNAs can exert dual biological effects on cancer through multiple regulatory mechanisms (Li XZ et al., 2021; di Fazio and Gullerova, 2023). For example, tRF5-Glu has been reported to inhibit the expression of breast cancer anti-estrogen resistance 3 (BCAR3) by binding to its 3' untranslated region (UTR) position, thereby hindering the proliferation of breast cancer cells (Zhou et al., 2017). Luan et al. (2021) found that hypoxic colorectal cancer (CRC) cells induced the production of tRF-20-MEJB5Y13, which promoted CRC cell migration and invasion by mediating endothelial–mesenchymal transition (EMT). Taken together, tsRNAs have the potential to become tumor biomarkers for the dynamic monitoring of patients and provide new possibilities for targeted therapy.
In our previous study, we screened the highly expressed tRF-23-Q99P9P9NDD using high-throughput sequencing and verified its potential as a diagnostic marker for GC (Zhang et al., 2022). In the present work, we first indicated that tRF-23-Q99P9P9NDD could promote the proliferation, migration, and invasion of GC cells, and the target gene acyl-coenzyme A (CoA) dehydrogenase short/branched chain (ACADSB) could rescue the effect of tRF-23-Q99P9P9NDD on GC cells. ACADSB belongs to the acyl-CoA dehydrogenase family, which mainly catalyzes the dehydrogenation of acyl-CoA derivatives during the metabolism of fatty acids (FAs) and branch-chained amino acids (BCAAs) (Rozen et al., 1994). We have confirmed that downregulated ACADSB may promote the malignancy of GC by regulating lipid accumulation and ferroptosis. Overall, our findings indicated that tRF-23-Q99P9P9NDD promotes the progression of GC by targeting ACADSB and is expected to become a new therapeutic target for GC.
2. Results
2.1. Effects of tRF-23-Q99P9P9NDD on the proliferation, migration, and invasion of GC cells
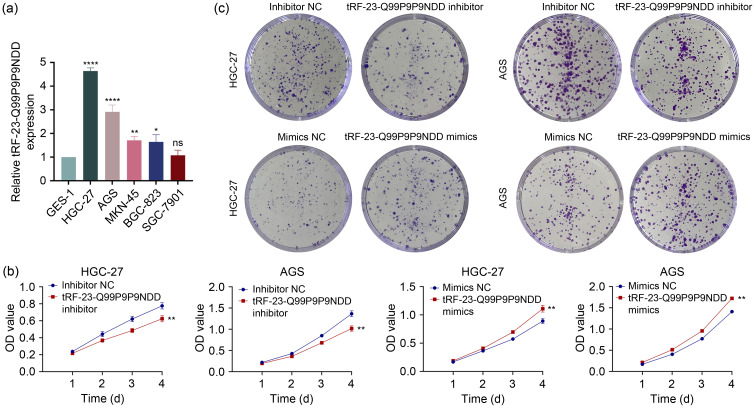
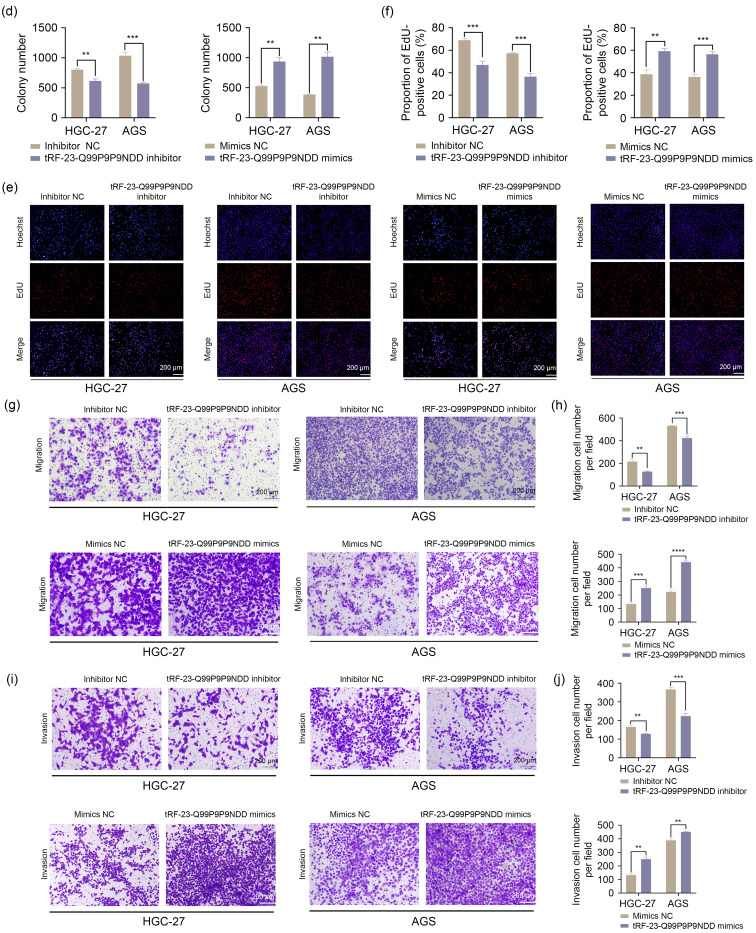
In order to explore whether tRF-23-Q99P9P9NDD affected the malignant progression of GC, we first detected the expression of tRF-23-Q99P9P9NDD in five GC cell lines by quantitative real-time polymerase chain reaction (qRT-PCR). We found that the expression level of tRF-23-Q99P9P9NDD was higher than that of GES-1 (Fig. 1a), especially in HGC-27 and AGS. Thus, we performed subsequent experiments after transfection with tRF-23-Q99P9P9NDD inhibitors and mimics in HGC-27 and AGS, and the transfection efficiency is shown in Fig. S1a. Cell counting kit-8 (CCK-8) and clone formation assays showed that the knockdown of tRF-23-Q99P9P9NDD inhibited GC cell viability and clone formation compared to the control (Figs. 1b‒1d). The 5-ethynyl-2'-deoxyuridine (EdU) experiments confirmed that GC cells treated with tRF-23-Q99P9P9NDD inhibitor reduced the level of DNA synthesis in cells. However, the above results showed the opposite trend after the overexpression of tRF-23-Q99P9P9NDD (Figs. 1e and 1f). In addition, we examined how tRF-23-Q99P9P9NDD affected the motility of GC cells by transwell assays. The results revealed that the knockdown of tRF-23-Q99P9P9NDD significantly inhibited the migration and invasion of GC cells. On the contrary, the overexpression of tRF-23-Q99P9P9NDD accelerated the migration and invasion of GC cells (Figs. 1g‒1j). These results indicated that tRF-23-Q99P9P9NDD could promote the proliferation, migration, and invasion of GC cells.
Fig. 1. Effects of tRF-23-Q99P9P9NDD on the proliferation, migration, and invasion of GC cells. (a) The expression level of tRF-23-Q99P9P9NDD in GC cells was detected by qRT-PCR. (b‒f) The effects of transfection with tRF-23-Q99P9P9NDD inhibitor and mimics on the proliferation of HGC-27 and AGS were evaluated by CCK-8, cell colony formation assay, and EdU assay. (g‒j) The effects of transfection with tRF-23-Q99P9P9NDD inhibitor and mimics on the migration and invasion ability of HGC-27 and AGS were evaluated by transwell assay. Data were expressed as mean±standard deviation (SD), n=3. ** P<0.01, *** P<0.001. GC: gastric cancer; NC: negative control; qRT-PCR: quantitative real-time polymerase chain reaction; CCK-8: cell counting kit-8; EdU: 5-ethynyl-2'-deoxyuridine; OD:optical density.
2.2. Effect of tRF-23-Q99P9P9NDD on the expression of ACADSB in GC cells
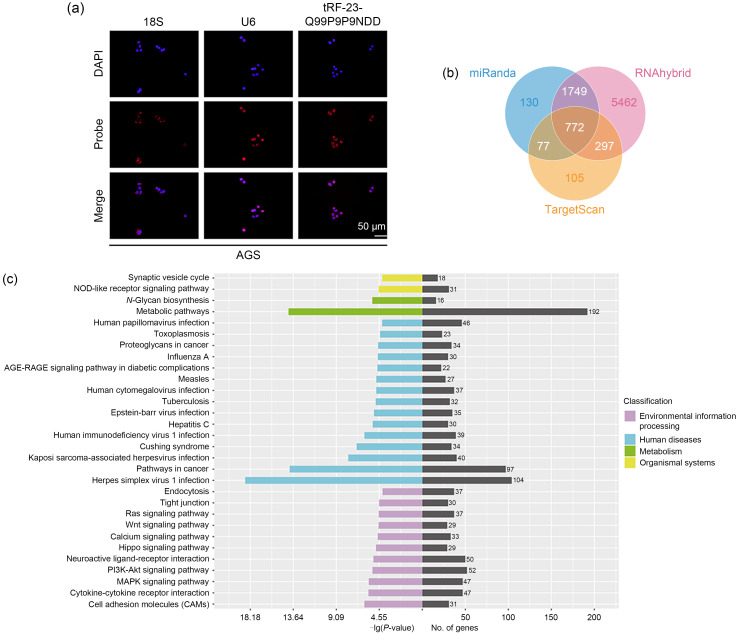
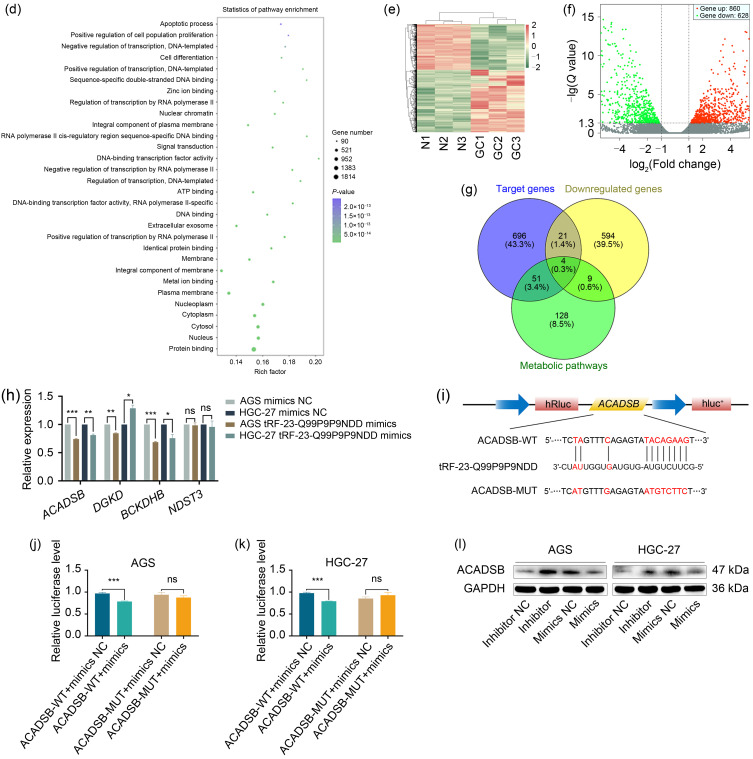
In order to fully investigate the mechanism of tRF-23-Q99P9P9NDD affecting GC progression, fluorescence in situ hybridization (FISH) experiment was first performed in AGS to confirm that tRF-23-Q99P9P9NDD was mainly distributed throughout the nucleoplasm (Fig. 2a). Since tsRNAs are similar to miRNAs in size and structure, they can also inhibit gene expression through RNA interference (RNAi) (Han et al., 2022). We used the TargetScan (https://www.targetscan.org/vert_80), miRanda (http://www.microrna.org/microrna/home.do), and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) databases to predict target genes that have binding sites with tRF-23-Q99P9P9NDD at the 3' UTR position (Fig. 2b). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that herpes simplex virus 1 infection, metabolic pathways, and pathways in cancer presented significant changes (Fig. 2c). Gene Ontology (GO) analysis indicated that these target genes were significantly enriched in protein binding, nucleus, cytosol, and cytoplasm (Fig. 2d). In addition, we performed high-throughput sequencing of the previous three pairs of GC tissues and their paired non-tumor adjacent tissues, and visualized the differentially expressed genes by heatmaps and volcano plots (Figs. 2e and 2f). Finally, we combined the predicted target genes of tRF-23-Q99P9P9NDD with the metabolic pathway in the KEGG pathway (pathway ID: hsa01100) and the downregulated messenger RNAs (mRNAs) in the sequencing to screen out four candidate target genes (ACADSB, diacylglycerol kinase delta(DGKD),branched chain keto acid dehydrogenase E1, subunit beta(BCKDHB), and N-deacetylase and N-sulfotransferase 3 (NDST3)) (Fig. 2g). The expression levels of these target genes were detected after transfecting mimics of tRF-23-Q99P9P9NDD in HGC-27 and AGS, and the most significantly downregulated ACADSB was selected as the target gene of tRF-23-Q99P9P9NDD for further study (Fig. 2h).
Fig. 2. Effects of tRF-23-Q99P9P9NDD on the expression of ACADSB in GC cells. (a) The localization of tRF-23-Q99P9P9NDD in GC cells was determined by FISH experiments. (b) Prediction of target genes for tRF-23-Q99P9P9NDD based on the TargetScan, miRanda, and RNAhybrid databases. (c) KEGG biological pathway functional enrichment analysis of the target genes of tRF-23-Q99P9P9NDD. (d) GO functional enrichment analysis of the target genes of tRF-23-Q99P9P9NDD. (e, f) Heatmap and volcano plot analysis of genes differentially expressed in three GC tissues (GC1‒GC3) and paired non-tumor adjacent tissues (N1‒N3). (g) The prediction of target genes, metabolic pathways, and sequencing of downregulated genes were combined to identify target genes for tRF-23-Q99P9P9NDD. (h) The expression of four target genes after transfection with tRF-23-Q99P9P9NDD inhibitor and mimics by qRT-PCR. (i) Schematic diagram of the binding site between WT ACADSB and tRF-23-Q99P9P9NDD. (j, k) The binding of tRF-23-Q99P9P9NDD to ACADSB was verified by dual fluorescein-free reporter gene assay in AGS and HGC-27. (l) The expression of ACADSB after transfection with tRF-23-Q99P9P9NDD inhibitor and mimics by western blot. Data were expressed as mean±standard deviation (SD), n=3. * P<0.05, ** P<0.01, *** P<0.001. ACADSB: acyl-coenzyme A dehydrogenase short/branched chain; GC: gastric cancer; FISH: fluorescence in situ hybridization; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; qRT-PCR: quantitative real-time polymerase chain reaction; WT: wild-type; MUT: mutant; NC: negative control; ns: not significant; DAPI: 4',6-diamidino-2-phenylindole; DGKD: diacylglycerol kinase delta; BCKDHB: branched chain keto acid dehydrogenase E1, subunit beta; NDST3: N-deacetylase and N-sulfotransferase 3; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
In order to verify whether there was a direct binding relationship between tRF-23-Q99P9P9NDD and ACADSB, we co-transfected luciferase reporter vectors containing ACADSB 3' UTR wild type (WT) or mutant (MUT) (Fig. 2i) with tRF-23-Q99P9P9NDD mimics or negative control (NC) in AGS and HGC-27. As shown in Figs. 2j and 2k, compared with the NC group, mimics of tRF-23-Q99P9P9NDD inhibited the luciferase activity of the ACADSB-WT group. However, the tRF-23-Q99P9P9NDD mimics or NC group did not affect the luciferase activity in cells transfected with ACADSB-MUT. The above results confirmed that tRF-23-Q99P9P9NDD had binding sites with the 3' UTR region of ACADSB. Next, we established that the knockdown of tRF-23-Q99P9P9NDD increased the protein level of ACADSB in GC cells, and the overexpression of tRF-23-Q99P9P9NDD showed the opposite trend (Fig. 2l). These results suggested that tRF-23-Q99P9P9NDD targeted ACADSB and inhibited its expression level.
2.3. Downregulated ACADSB in GC
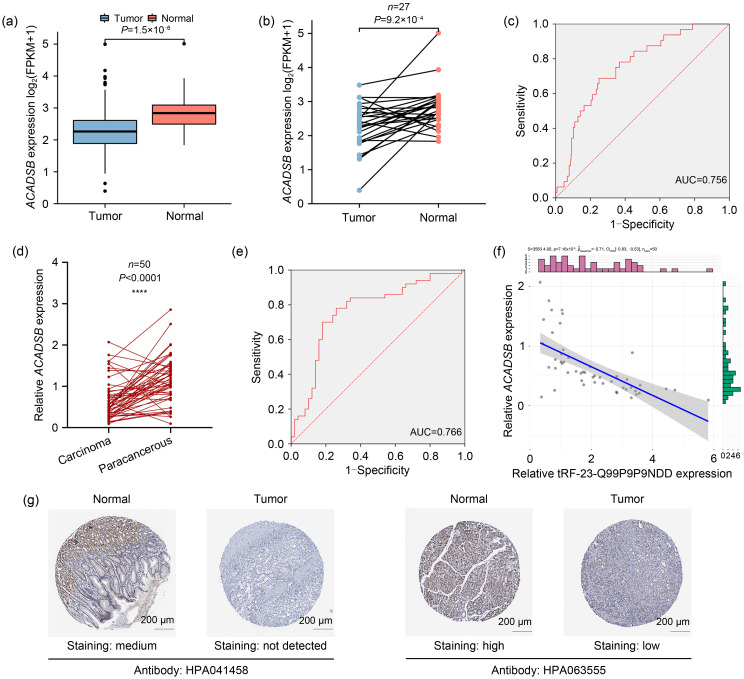
By searching The Cancer Genome Atlas (TCGA) stomach adenocarcinoma (STAD) database, we found that the expression level of ACADSB in GC tissues was significantly lower than that in adjacent normal tissues (Fig. 3a), and its expression in paired GC tissues was also lower than that in non-tumor tissues (Fig. 3b). Receiver operating characteristic (ROC) analysis showed that the area under the curve (AUC) of ACADSB in the TCGA STAD database was 0.756 (Fig. 3c). Next, we detected the level of ACADSB in fifty pairs of GC tissues by qRT-PCR, and the results showed that it was lowly expressed in GC (Fig. 3d). In addition, ROC analysis revealed that the AUC of ACADSB was 0.766 (Fig. 3e), demonstrating its good diagnostic efficacy for GC, consistent with the database findings. Spearman correlation analysis was performed to find that tRF-23-Q99P9P9NDD and ACADSB were negatively correlated in GC tissues (Fig. 3f), which proved that tRF-23-Q99P9P9NDD might have a regulatory effect on ACADSB. In addition, according to the immunohistochemical staining results of two different ACADSB antibodies in the Human Protein Atlas (HPA) database, the staining was lighter in GC tissues than in normal gastric tissues, indicating that ACADSB protein expression was relatively low in GC tissues (Fig. 3g).
Fig. 3. Downregulated ACADSB in GC. (a) Expression level of ACADSB in TCGA STAD database (tumor: n=373; normal: n=32). (b) Expression level of ACADSB in paired GC samples in TCGA STAD database. (c) ROC curve of ACADSB in TCGA STAD database. (d) Expression level of ACADSB in GC tissues. (e) ROC curve of ACADSB in GC tissues. (f) Correlation between ACADSB and tRF-23-Q99P9P9NDD in GC tissues. (g) Comparison of immunohistochemistry images of ACADSB between GC and normal gastric tissues based on the HPA database. ACADSB: acyl-coenzyme A dehydrogenase short/branched chain; GC: gastric cancer; TCGA: The Cancer Genome Atlas; STAD: stomach adenocarcinoma; ROC: receiver operating characteristic; AUC: area under the curve; HPA: Human Protein Atlas; FPKM: fragments per kilobase per million.
2.4. Effects of ACADSB on the proliferation, migration, and invasion of GC cells
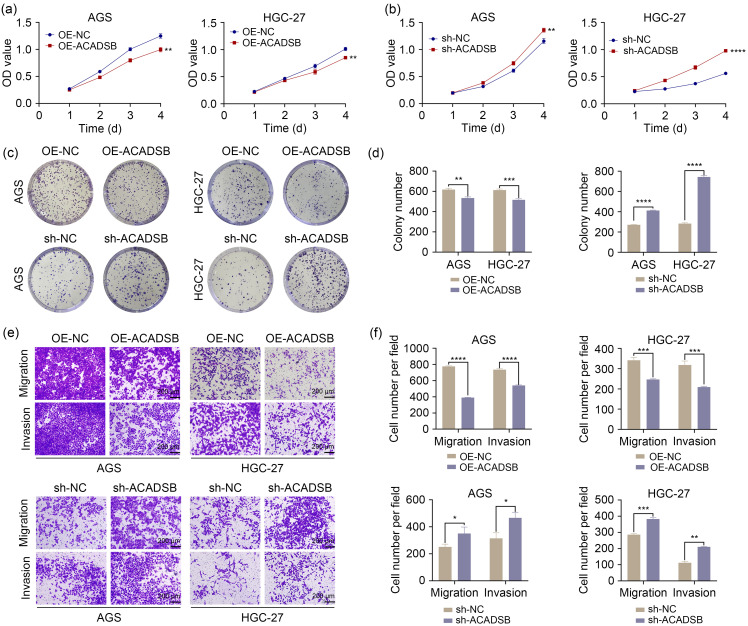
Next, we transfected plasmids overexpressing and knocking down ACADSB in AGS and HGC-27 and detected the transfection efficiency through qRT-PCR and western blot (Figs. S1b and S1c). As shown in Fig. 4, the overexpression of ACADSB inhibited the proliferation, migration, and invasion of GC cells. However, the knockdown of ACADSB promoted the progression of GC, similar to the effect of tRF-23-Q99P9P9NDD mimics.
Fig. 4. Effects of ACADSB on the proliferation, migration, and invasion of GC cells. (a, b) CCK-8 assays in AGS and HGC-27 after overexpression (a) or knockdown (b) of ACADSB. (c, d) Cell colony formation assays in AGS and HGC-27 after overexpression and knockdown of ACADSB. (e, f) Transwell assays in AGS and HGC-27 after overexpression and knockdown of ACADSB. Data were expressed as mean±standard deviation (SD), n=3. * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. ACADSB: acyl-coenzyme A dehydrogenase short/branched chain; CCK-8: cell counting kit-8; GC: gastric cancer; OE: overexpression; NC: negative control; sh: short hairpin RNA; OD:optical density.
2.5. Rescue effect of ACADSB against tRF-23-Q99P9P9NDD on GC cells
We conducted rescue experiments to further explore whether tRF-23-Q99P9P9NDD could promote the malignant activity of GC cells by regulating ACADSB. As shown in Fig. 5, the overexpression of ACADSB reversed the enhanced ability of proliferation, migration, and invasion induced by tRF-23-Q99P9P9NDD. In addition, the knockdown of ACADSB restored the damaging effect of the tRF-23-Q99P9P9NDD inhibitor on GC cells. These results suggested that tRF-23-Q99P9P9NDD may promote the proliferation, migration, and invasion of GC cells by inhibiting the target gene ACADSB.
Fig. 5. Rescue effect of ACADSB against tRF-23-Q99P9P9NDD on GC cells. Transfection of OE-NC+mimics NC, OE-NC+mimics, OE-ACADSB+mimics NC, and OE-ACADSB+mimics into AGS; and transfection of sh-NC+inhibitor NC, sh-NC+inhibitor, sh-ACADSB+inhibitor NC, and sh-ACADSB+inhibitor into HGC-27 cells. (a‒d) The effects on the proliferative capacity of AGS and HGC-27 under different conditions were evaluated by CCK-8 and cell colony formation assays. (e‒h) The effects on the migration and invasion ability of AGS and HGC-27 under different conditions were evaluated by transwell migration and invasion assays. Data were expressed as mean±standard deviation (SD), n=3. ** P<0.01, *** P<0.001, **** P<0.0001. ACADSB: acyl-coenzyme A dehydrogenase short/branched chain; GC: gastric cancer; OE: overexpression; NC: negative control; sh: short hairpin RNA; CCK-8: cell counting kit-8; OD:optical density.
2.6. Function and pathway prediction of ACADSB enrichment in GC
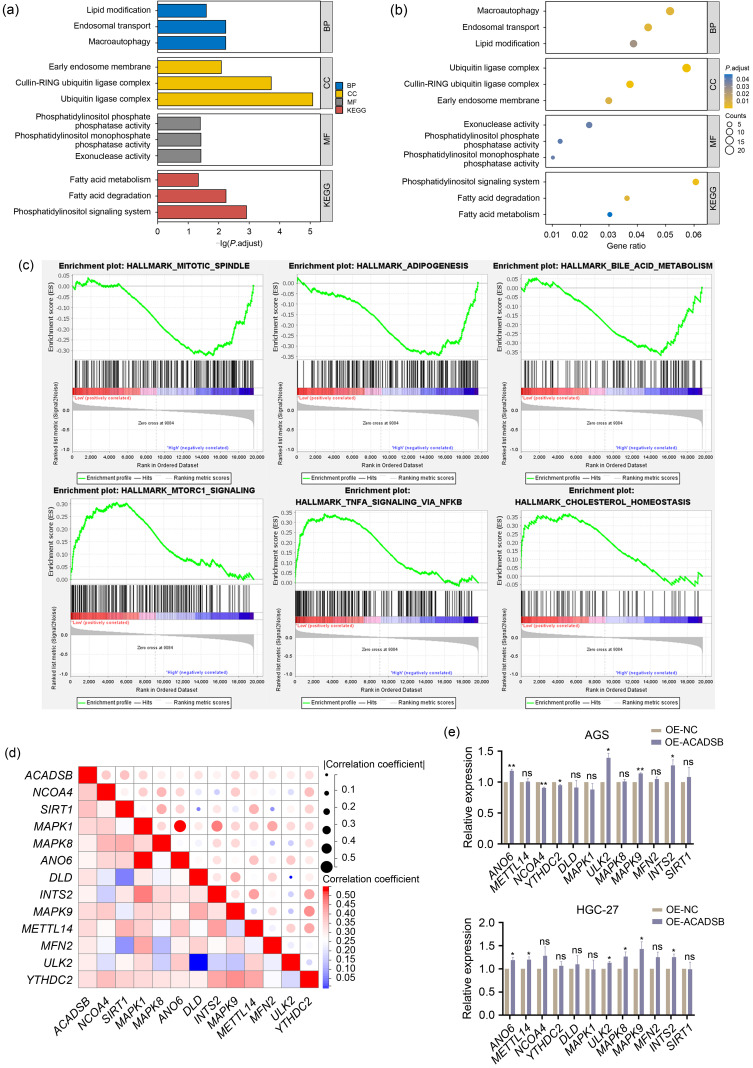
In order to explore the mechanism of tRF-23-Q99P9P9NDD targeting ACADSB to promote GC progression, we performed GO and KEGG analyses on 431 genes with a correlation greater than 0.3 between their expression levels in the TCGA STAD database and those of ACADSB. The GO analysis showed significant lipid modification, endosomal transport, and macroautophagy enrichment in the biological process (BP). Ubiquitin ligase complex, cullin-RING ubiquitin ligase complex, and early endosome membrane were significantly enriched in cellular component (CC). The main differences in molecular function (MF) included exonuclease activity, phosphatidylinositol monophosphate phosphatase activity, and phosphatidylinositol phosphate phosphatase activity. KEGG analysis showed that these genes were enriched in the phosphatidylinositol signaling system, FA degradation, and FA metabolism pathways (Figs. 6a and 6b). The Gene Set Enrichment Analysis (GSEA) results showed that the ACADSB high-expression phenotype was significantly enriched in MITOTIC_SPINDLE, ADIPOGENESIS, and BILE_ACID_METABOLISM, while the ACADSB low-expression phenotype was in MTORC1_SIGNALING, TNFA_SIGNALING_VIA_NFKB, and CHOLESTEROL_HOMEOSTASIS (Fig. 6c). The above results indicated that ACADSB might be associated with lipid metabolism in GC.
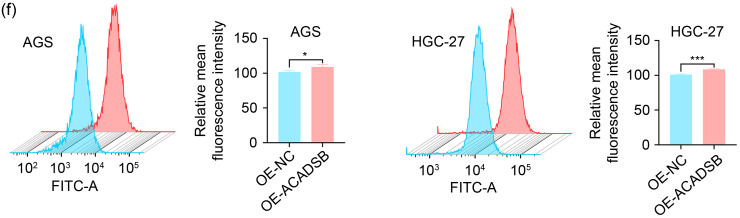
Fig. 6. Function and pathway prediction of ACADSB enrichment in GC and the relationship between ACADSB and ferroptosis driver genes. (a, b) GO and KEGG analyses on 431 ACADSB positively related genes. (c) Enriched pathways in the high and low ACADSB groups based on GSEA. (d) The expression level of ACADSB in the TCGA STAD database is positively correlated with 12 ferroptosis driver genes. (e) Correlation between ACADSB and 12 ferroptosis genes at the transcriptional level. (f) Changes in ROS levels after the overexpression of ACADSB were detected by flow cytometry. ACADSB: acyl-coenzyme A dehydrogenase short/branched chain; GC: gastric cancer; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; GSEA: Gene Set Enrichment Analysis; BP: biological process; CC: cellular component; MF: molecular function; TCGA: The Cancer Genome Atlas; STAD: stomach adenocarcinoma; ROS: reactive oxygen species; NC: negative control; OE: overexpression; FITC: fluorescein isothiocyanate.
2.7. Correlation of ACADSB with ferroptosis
Lipid metabolism is closely linked to ferroptosis (Stockwell et al., 2017). Previous studies found that ACADSB could inhibit the malignancy of CRC by regulating ferroptosis in CRC cells (Lu et al., 2020; Zhou and Bao, 2020). The FerrDb database (http://www.zhounan.org/ferrdb) also showed that ACADSB belonged to the ferroptosis driver genes. This study defined 227 genes as ferroptosis driver genes based on the FerrDb database. According to correlation analysis, the expression level of ACADSB in the TCGA STAD database was positively correlated with 12 ferroptosis genes, including nuclear receptor coactivator 4 (NCOA4), sirtuin 1 (SIRT1), mitogen-activated protein kinase 1 (MAPK1), and MAPK8 with r>0.3 (Fig. 6d). Next, we detected the expression of these 12 ferroptosis genes after ACADSB overexpression in AGS and HGC-27 cells by qRT-PCR. Compared with the control group, anoctamin 6 (ANO6), Unc-51-like autophagy-activating kinase 2 (ULK2), MAPK9, and integrator complex subunit 2 (INTS2) were significantly upregulated after ACADSB overexpression (Fig. 6e). We further confirmed by flow cytometry that reactive oxygen species (ROS) levels were increased after the overexpression of ACADSB (Fig. 6f). The above results suggested that ACADSB might act as a driver gene of ferroptosis and simultaneously regulate other ferroptosis driver genes to promote ferroptosis, thus inhibiting the progression of GC.
3. Discussion
Due to the high tumor heterogeneity and late diagnosis period, the mortality rate of GC continues to be high despite the significant treatment advances (Lai et al., 2020; Sung et al., 2021). Therefore, it is crucial to find biomarkers that can provide assistance in the early diagnosis of GC and explore the mechanisms that regulate GC progression. As a new type of ncRNAs, tsRNAs are widely distributed in the body and have tissue or developmental stage-specific characteristics (Hsieh et al., 2009; Torres et al., 2019). Researchers have found that dysregulated tsRNAs affect cancer progression through transcriptional gene silencing, post-transcriptional gene silencing, reverse transcription regulation, and translation regulation (Li J et al., 2021). For example, tRF3E inhibits the growth of breast cancer cells by forming a complex with nucleolin to destroy the inhibitory effect of nucleolin on the translation of protein 53 (p53) (Falconi et al., 2019). After binding to the target sites of ribosomal protein S28 (RPS28) mRNA, LeuCAG3'tsRNA enhances translation by unfolding its secondary hairpin structure. Thus, the inhibition of LeuCAG3'tsRNA may lead to blocked translation of RPS28, inducing apoptosis and acting as a target for hepatocellular carcinoma therapy (Kim et al., 2017). Therefore, tsRNAs have the potential to become GC diagnostic markers and therapeutic targets, whereas the exact mechanism remains to be further investigated.
In this study, we first confirmed that tRF-23-Q99P9P9NDD could promote the proliferation, migration, and invasion of GC cells. Next, we screened out the most significantly downregulated ACADSB as the target gene and found that ACADSB could rescue the effect of tRF-23-Q99P9P9NDD on GC cells. Bioinformatics analysis suggested that downregulated ACADSB might promote GC progression by inhibiting ferroptosis and promoting lipid accumulation. In summary, this study proposes that tRF-23-Q99P9P9NDD may be involved in the lipid metabolism and ferroptosis of GC by targeting ACADSB, thereby promoting the progression of GC.
Mechanistically, because tRFs are similar to miRNAs, they can bind to the 3' UTR region of target mRNAs to inhibit gene expression (Haussecker et al., 2010). For example, tRF/miR-1280 impedes CRC cell metastasis by targeting the 3' UTR region of Notch ligand Jagged-2 (JAG2) mRNA (Huang et al., 2017). In the present study, we found that tRF-23-Q99P9P9NDD could negatively regulate the target gene ACADSB, and the mRNA and protein levels of ACADSB decreased significantly after transfection with tRF-23-Q99P9P9NDD mimics. Thus, we used the dual luciferase reporter gene assay to confirm that tRF-23-Q99P9P9ND can bind to the 3' UTR of ACADSB, similar to miRNA-mediated target gene silencing.
The downregulation of ACADSB has been reported in diverse cancers (Ericksen et al., 2019; Lu et al., 2020) but has not been studied in GC. Our study demonstrated that ACADSB had good diagnostic performance in GC. GO, KEGG, and GSEA analyses were performed to show that ACADSB might be related to lipid metabolism. In addition, ACADSB-related genes were enriched in FA degradation and metabolism. Therefore, downregulated ACADSB in GC could lead to lipid accumulation by inhibiting FA catabolism (Rozen et al., 1994; Carracedo et al., 2013; Qu et al., 2020). Interestingly, lipid metabolism is related to ferroptosis, a form of iron-dependent cell death caused by the accumulation of lipid hydroperoxides (Stockwell et al., 2017; Qu et al., 2022). In clear cell renal carcinoma (ccRCC), lowly expressed ACADSB accelerates the progression of ccRCC by inhibiting FA and BCAA catabolism and ferroptosis (Liu et al., 2022). Based on our results, ACADSB had a positive correlation with some ferroptosis-driven genes and the ROS levels increased after the overexpression of ACADSB. Therefore, we revealed that downregulated ACADSB could contribute to the malignancy of GC by promoting lipid accumulation and inhibiting ferroptosis.
However, our study has some limitations. For example, the regulatory relationship between tRF-23-Q99P9P9NDD and ACADSB in GC remains to be further investigated. In addition, the overall effect on tRF-23-Q99P9NDD and other collaborative pathways needs to be further studied. Finally, experiments are needed to further verify the relationship between ACADSB and lipid metabolism and ferroptosis in GC. In summary, we demonstrated for the first time that tRF-23-Q99P9P9NDD may affect lipid metabolism and GC ferroptosis by targeting ACADSB, thereby promoting the proliferation, migration, and invasion of GC cells. This study provides a theoretical and experimental basis for tRF-23-Q99P9P9NDD to aid in the diagnosis and prognosis of GC and also presents a novel treatment target.
4. Conclusions
We indicated for the first time that tRF-23-Q99P9P9NDD can promote the proliferation, migration, and invasion of GC cells. Meanwhile, tRF-23-Q99P9P9NDD may be involved in the lipid metabolism and ferroptosis of GC by targeting ACADSB.
Materials and methods
Detailed methods are provided in the electronic supplementary materials of this paper.
Supplementary information
Acknowledgments
We appreciate all the patients implicated in this study and all those who contributed to it. This work was supported by the National Natural Science Foundation of China (Nos. 82272411 and 82072363), the Jiangsu Provincial Medical Key Discipline (Laboratory) (No. ZDXK202240), and the Science and Technology Project of Jiangsu Province (No. BE2023741), China.
Author contributions
Yu ZHANG and Xinliang GU performed study design, data collection, and analysis. Yu ZHANG wrote the draft of the manuscript. Xinliang GU supervised and lead the planning and execution of research activities. Yang LI took part in the experiment. Xun LI revised the manuscript. Yuejiao HUANG and Shaoqing JU provided guidance for the study. All authors have read and approved this manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Yu ZHANG, Xinliang GU, Yang LI, Xun LI, Yuejiao HUANG, and Shaoqing JU declare that they have no conflict of interest.
The study was approved by the Affiliated Hospital of Nantong University (ethical review report number: 2018-L055) and performed in accordance with the Helsinki Declaration of 1975, as revised in 2013. Informed consent was obtained from all patients for being included in the study.
Data availability statement
The data that support the findings of the current study are available from the corresponding author upon reasonable request.
References
- Carracedo A, Cantley LC, Pandolfi PP, 2013. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer, 13(4): 227-232. 10.1038/nrc3483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P, 2013. Gastric cancer: overview. Gastroenterol Clin North Am, 42(2): 211-217. 10.1016/j.gtc.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Fazio A, Gullerova M, 2023. An old friend with a new face: tRNA-derived small RNAs with big regulatory potential in cancer biology. Br J Cancer, 128(9): 1625-1635. 10.1038/s41416-023-02191-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericksen RE, Lim SL, Mcdonnell E, et al. , 2019. Loss of BCAA catabolism during carcinogenesis enhances mTORC1 activity and promotes tumor development and progression. Cell Metab, 29(5): 1151-1165.e6. 10.1016/j.cmet.2018.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconi M, Giangrossi M, Zabaleta ME, et al. , 2019. A novel 3'-tRNAGlu-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J, 33(12): 13228-13240. 10.1096/fj.201900382RR [DOI] [PubMed] [Google Scholar]
- Han Y, Peng YH, Liu SS, et al. , 2022. tRF3008A suppresses the progression and metastasis of colorectal cancer by destabilizing FOXK1 in an AGO-dependent manner. J Exp Clin Cancer Res, 41: 32. 10.1186/s13046-021-02190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haussecker D, Huang Y, Lau A, et al. , 2010. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA, 16(4): 673-695. 10.1261/rna.2000810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh LC, Lin SI, Shih ACC, et al. , 2009. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol, 151(4): 2120-2132. 10.1104/pp.109.147280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang BQ, Yang HP, Cheng XX, et al. , 2017. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res, 77(12): 3194-3206. 10.1158/0008-5472.Can-16-3146 [DOI] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang SC, et al. , 2017. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature, 552(7683): 57-62. 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Kuscu C, Dutta A, 2016. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci, 41(8): 679-689. 10.1016/j.tibs.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SW, Kuo YH, Liao KF, 2020. Statin therapy and gastric cancer death. Postgrad Med J, 96(1133): 178. 10.1136/postgradmedj-2019-136994 [DOI] [PubMed] [Google Scholar]
- Li J, Zhu L, Cheng J, et al. , 2021. Transfer RNA-derived small RNA: a rising star in oncology. Semin Cancer Biol, 75: 29-37. 10.1016/j.semcancer.2021.05.024 [DOI] [PubMed] [Google Scholar]
- Li XZ, Liu XY, Zhao DZ, et al. , 2021. tRNA-derived small RNAs: novel regulators of cancer hallmarks and targets of clinical application. Cell Death Discov, 7: 249. 10.1038/s41420-021-00647-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu BW, Cao JL, Wang XY, et al. , 2021. Deciphering the tRNA-derived small RNAs: origin, development, and future. Cell Death Dis, 13: 24. 10.1038/s41419-021-04472-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XH, Zhang WY, Wang HR, et al. , 2022. Decreased expression of ACADSB predicts poor prognosis in clear cell renal cell carcinoma. Front Oncol, 11: 762629. 10.3389/fonc.2021.762629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Yang ZY, Xia QY, et al. , 2020. ACADSB regulates ferroptosis and affects the migration, invasion, and proliferation of colorectal cancer cells. Cell Biol Int, 44(11): 2334-2343. 10.1002/cbin.11443 [DOI] [PubMed] [Google Scholar]
- Luan N, Mu YL, Mu JY, et al. , 2021. Dicer1 promotes colon cancer cell invasion and migration through modulation of tRF-20-MEJB5Y13 expression under hypoxia. Front Genet, 12: 638244. 10.3389/fgene.2021.638244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EJ, Kim TH, 2018. Fine-tuning of gene expression by tRNA-derived fragments during abiotic stress signal transduction. Int J Mol Sci, 19(2): 518. 10.3390/ijms19020518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L, He XY, Tang Q, et al. , 2022. Iron metabolism, ferroptosis, and lncRNA in cancer: knowns and unknowns. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 23(10): 844-862. 10.1631/jzus.B2200194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu YY, Zhao R, Zhang HL, et al. , 2020. Inactivation of the AMPK-GATA3-ECHS1 pathway induces fatty acid synthesis that promotes clear cell renal cell carcinoma growth. Cancer Res, 80(2): 319-333. 10.1158/0008-5472.Can-19-1023 [DOI] [PubMed] [Google Scholar]
- Rozen R, Vockley J, Zhou LB, et al. , 1994. Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family. Genomics, 24(2): 280-287. 10.1006/geno.1994.1617 [DOI] [PubMed] [Google Scholar]
- Shen L, Shan YS, Hu HM, et al. , 2013. Management of gastric cancer in Asia: resource-stratified guidelines. Lancet Oncol, 14(12): e535-e547. 10.1016/s1470-2045(13)70436-4 [DOI] [PubMed] [Google Scholar]
- Stockwell BR, Angeli JPF, Bayir H, et al. , 2017. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell, 171(2): 273-285. 10.1016/j.cell.2017.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, et al. , 2021. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 71(3): 209-249. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- Tao EW, Cheng WY, Li WL, et al. , 2020. tiRNAs: a novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol, 235(2): 683-690. 10.1002/jcp.29057 [DOI] [PubMed] [Google Scholar]
- Torres AG, Reina O, Stephan-Otto Attolini C, et al. , 2019. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc Natl Acad Sci USA, 116(17): 8451-8456. 10.1073/pnas.1821120116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen JT, Huang ZH, Li QH, et al. , 2021. Research progress on the tsRNA classification, function, and application in gynecological malignant tumors. Cell Death Discov, 7: 388. 10.1038/s41420-021-00789-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DD, Qiao DQ, Lei YL, et al. , 2022. Transfer RNA-derived small RNAs (tsRNAs): versatile regulators in cancer. Cancer Lett, 546: 215842. 10.1016/j.canlet.2022.215842 [DOI] [PubMed] [Google Scholar]
- Yang M, Mo YZ, Ren DX, et al. , 2023. Transfer RNA-derived small RNAs in tumor microenvironment. Mol Cancer, 22: 32. 10.1186/s12943-023-01742-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Gu XL, Qin XY, et al. , 2022. Evaluation of serum tRF-23-Q99P9P9NDD as a potential biomarker for the clinical diagnosis of gastric cancer. Mol Med, 28: 63. 10.1186/s10020-022-00491-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Diebel KW, Holy J, et al. , 2017. A tRNA fragment, tRF5-Glu, regulates BCAR3 expression and proliferation in ovarian cancer cells. Oncotarget, 8(56): 95377-95391. 10.18632/oncotarget.20709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou N, Bao JK, 2020. FerrDb: a manually curated resource for regulators and markers of ferroptosis and ferroptosis-disease associations. Database, 2020: baaa021. 10.1093/database/baaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of the current study are available from the corresponding author upon reasonable request.