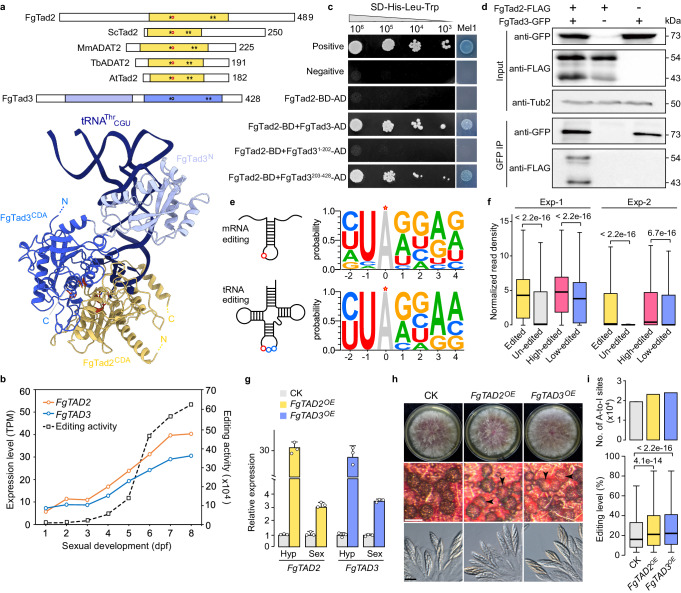
Fig. 1. A-to-I editing of tRNA and mRNA by the FgTad2-FgTad3 complex.
a Domain structures of FgTad2 and FgTad3, along with the AlphaFold model depicting FgTad2-FgTad3 bound to tRNA. Orange, Tad2 CDA; blue, FgTad3CDA; sky blue, FgTad3 N-terminal domain (FgTad3N); Asterisk (*), Zn2+ coordinating residue; red circle, active-site glutamate; black circle, pseudo-active-site valine. For comparison, domain structures of Tad2/ADAT2 from Saccharomyces cerevisiae (Sc), Mus musculus (Mm), Trypanosoma brucei (Tb), and Arabidopsis thaliana (At) are shown. b Expression levels of FgTAD2 and FgTAD3, as well as editing activity (summing the editing levels of all editing sites) assessed by RNA-seq. c Y2H assays for the interaction between FgTad2 (BD-bait) and the full-length protein, N-terminal (1-202 aa), or C-terminal (203-428 aa) region of FgTad3 (AD-prey). Photos show growth on SD-His-Leu-Trp plates at indicated concentrations, accompanied by Mel1 α-galactosidase activity. d Co-IP assays for the interaction between FgTad2 and FgTad3. Total proteins (input) isolated from transformants expressing FgTad2-3×FLAG and/or FgTad3-GFP and proteins eluted from anti-GFP affinity beads (GFP IP) were detected with anti-FLAG and anti-GFP antibodies. e WebLogo showing base preferences for tRNA and mRNA editing in F. graminearum. Editing sites are indicated by red circles or stars. f Boxplots of normalized read densities for edited (n = 6795), un-edited (n = 4641), high-edited (n = 3371), and low-edited (n = 3390) genes from two independent experiments (Exp-1 and Exp-2) of FgTad2-FLAG RIP-Seq. g Expression levels of FgTAD2 and FgTAD3 in hyphae (Hyp) and perithecia (Sex) of the FG1G36140 locus deletion strain (CK) and overexpressing transformants (OE) assayed by qRT-PCR. Data are presented as mean ± SD, n = 3 independent replicates. h Morphology of colonies, perithecia, and asci/ascospores of marked strains. Arrowheads indicate transparent perithecia. White bar = 0.2 mm; black bar = 20 μm. i Number of A-to-I editing sites and editing levels of shared sites (n = 22,356) in 7-dpf perithecia of marked strains. For the bar graph, data are presented as mean of two independent replicates. For (f, i), boxplots indicate median (middle line), 25th, 75th percentile (box), and 1.58x interquartile range (whisker). P values are from two-tailed Wilcoxon rank sum tests. Source data are provided as a Source Data file.