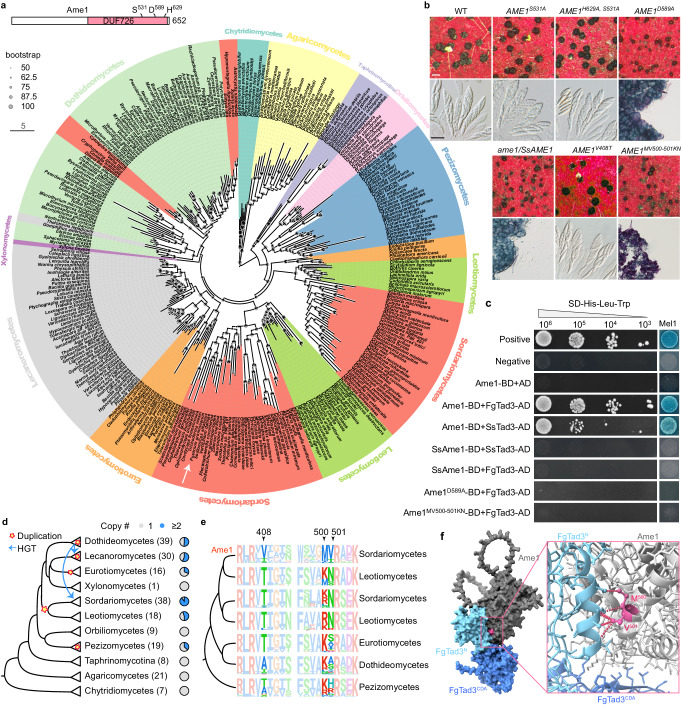
Fig. 5. Evolutionary origin of Ame1 and its key residues for mRNA editing in Sordariomycetes.
a The protein domain of Ame1 and a Maximum Likelihood phylogenetic tree of Ame1 orthologs from different fungal lineages. The putative catalytic triad (S-D-H) active sites are indicated. The white arrow indicates the position of AME1 in the tree. b Morphology of perithecia and asci/ascospores of different AME1 mutants. White bar = 0.2 mm; black bar = 20 μm. c Y2H assays for the interaction among Ame1, Ame1 mutants, FgTad3, and their orthologs SsAme1 and SsTad3 from Sclerotinia sclerotiorum. d The dendrogram of different fungal taxa showing evolutionary events related to Ame1 orthologs. The number of species in each taxon examined is indicated in the bracket. Each pie chart shows the proportion of fungal species with 1 or ≥ 2 copies of AME1 orthologs derived from gene duplication and/or horizontal gene transfer events. The putative origin of gene duplication and horizontal gene transfer events are indicated in the dendrogram. e WebLogo illustrating the conservation of amino acid residues within different fungal taxa at three putative sites responsible for the mRNA editing ability of Ame1. f The protein model showing the direct contact between the MV500-501 residues of Ame1 and the interface residues of FgTad3.