Significance
Alternative lengthening of telomeres (ALT) is a mechanism by which telomere length changes significantly in the absence of telomerase, the enzyme that typically maintains telomere length. ALT is a characteristic of 15% of cancers. A hallmark of ALT-positive cancers is the presence of C-circles, circular single-stranded DNAs enriched with C-rich sequences from telomeres. While these C-circles serve as a dependable diagnostic indicator, the definitive pathways of their formation are not known. This study addresses the underlying mechanism responsible for the generation of C-circles in ALT cancer cells, with a particular focus on C-circle generation from DNA replication of the lagging strand of telomeres, which require more complex steps for synthesis due to the limitations of the enzyme that replicates DNA.
Keywords: C-circle, BLM, POLD3, CST, break-induced replication
Abstract
Alternative lengthening of telomeres (ALT) is a telomere maintenance mechanism mediated by break-induced replication, evident in approximately 15% of human cancers. A characteristic feature of ALT cancers is the presence of C-circles, circular single-stranded telomeric DNAs composed of C-rich sequences. Despite the fact that extrachromosomal C-rich single-stranded DNAs (ssDNAs), including C-circles, are unique to ALT cells, their generation process remains undefined. Here, we introduce a method to detect single-stranded telomeric DNA, called 4SET (Strand-Specific Southern-blot for Single-stranded Extrachromosomal Telomeres) assay. Utilizing 4SET, we are able to capture C-rich single-stranded DNAs that are near 200 to 1500 nucleotides in size. Both linear C-rich ssDNAs and C-circles are abundant in the fractions of cytoplasm and nucleoplasm, which supports the idea that linear and circular C-rich ssDNAs are generated concurrently. We also found that C-rich ssDNAs originate during Okazaki fragment processing during lagging strand DNA synthesis. The generation of C-rich ssDNA requires CST-PP (CTC1/STN1/TEN1-PRIMASE-Polymerase alpha) complex-mediated priming of the C-strand DNA synthesis and subsequent excessive strand displacement of the C-rich strand mediated by the DNA Polymerase delta and the BLM helicase. Our work proposes a model for the generation of C-rich ssDNAs and C-circles during ALT-mediated telomere elongation.
Cancer cells acquire a mechanism to maintain telomere length in order to undergo neoplastic transformation. While most cancers activate telomerase, a reverse transcriptase which synthesizes telomere sequences, a subset of cancers acquire a mechanism called alternative lengthening of telomeres (ALT), which operates independently of telomerase and elongates telomeres through a DNA recombination-dependent process known as break-induced replication (BIR) (1–5). ALT cancers are characterized by ALT-associated biomarkers, including ALT-associated PML bodies (APBs) and extrachromosomal telomeric circular DNAs, such as C-circles (Cytosine-rich sequences (CCCTAA) containing single-stranded circular telomeric DNAs) and T-circles (t-loop sized circle; double-stranded circular telomeric DNAs) (6).
C-circles were initially found in ALT cancer and ALT-positive immortalized cell lines (7), making them valuable diagnostic markers for ALT cancer (8). These circular DNA molecules are present in low abundance and are mostly detectable through the use of phi29 polymerase (7). Furthermore, C-circles seem to preferentially form from the lagging strand (9, 10). The generation of C-circles in ALT cancer cells is thought to be related to replication stress experienced during S phases (11). Replication stress potentially activates the ALT pathway and leads to the formation of C-circles. However, the precise process by which C-circles are generated in ALT cancer cells remains unclear, necessitating further research to uncover the underlying mechanisms.
Apart from telomeric circular DNAs, ALT telomeres display various other structures, including T-bands, 5′ overhangs, and single-stranded telomeric DNAs (12, 13). T-bands and 5′ overhangs are predominantly observed in the chromatin fraction (13). The presence of 5′ overhangs is likely a consequence of telomere trimming, as they can be detected during T-circle generation processes (14).
Single-stranded telomeric DNAs, on the other hand, are extrachromosomal structures that are enriched in the cytoplasmic fraction obtained by Hirt Lysate (12, 13). However, the specific process through which extrachromosomal C-rich single-stranded telomere DNA generation occurs remains unclear and requires further investigation to understand its underlying mechanisms.
Results
4SET Assay, a Simple and Efficient Method for Detecting Single-Stranded Extrachromosomal Telomeric DNA.
To investigate the abundance of single-stranded telomeric DNA in ALT cancer, we have made modifications to existing Southern protocols that we named 4SET (Strand-Specific Southern-blot for Single-stranded Extrachromosomal Telomeres) (Fig. 1A). These modifications enhance sensitivity and optimize detection of single-stranded DNA. First, we optimized the genomic DNA extraction procedure for single-stranded DNA fragments (SI Appendix, Extended Methods). Compared to the conventional genomic DNA extraction kit (Qiagen DNeasy), our method is specifically optimized for the preparation of small-sized nucleic acids (SI Appendix, Fig. S1A). Loss of ssDNA using the Qiagen DNeasy extraction kit could be attributed to the use of columns where the small nucleic acids get trapped in the silica membrane and cannot be eluted. Second, we ran the genomic DNA on a low-percentage agarose gel and conducted the transfer onto a positively charged nylon membrane instead of using gel dry. This approach was chosen primarily to avoid the loss of small DNA fragments that can occur during the vacuum-based gel drying process. When the gel is dried using a vacuum, there is a risk of small DNA fragments being lost as the moisture is removed from below to dry the gel through the pressure. Last, we used highly sensitive strand-specific C-rich and G-rich telomere probe to determine strand specificity (15). This probe is optimal for detecting C-rich ssDNAs compared to probes based on end-labeling. In the whole DNA fraction, more ssDNA is observed with strand-specific probes (SI Appendix, Fig. S1B).
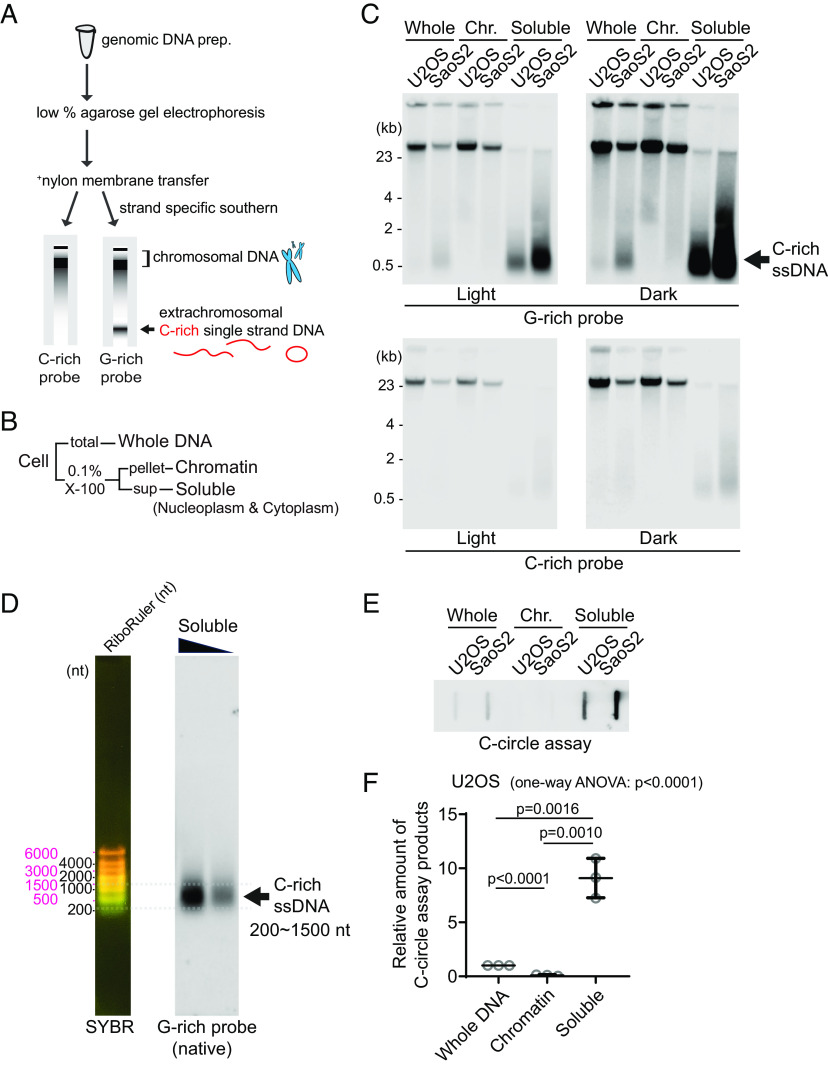
Fig. 1.
Detection of C-rich extrachromosomal telomeres. (A) Illustration of the Strand-specific Southern for Single-Stranded Extrachromosomal Telomeres (4SET) method. (Refer to the method for the details) (B) The 4SET method involves the fractionation of samples into Whole DNA fraction, Chromatin fraction, and Triton X-100 soluble fraction, achieved using 0.1% Triton X-100 extraction buffer. (C) 4SET assay for U2OS and SaoS2 cells after fractionation of samples into Whole, Chromatin (Chr.), and Soluble fractions. DNA amount: 600 ng. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. (D) 4SET assay with a nucleotide ladder (RiboRuler) to determine the size of C-rich single-stranded DNA in SaoS2 soluble fraction. G-rich probe was used for native gel to detect C-rich single-stranded telomeres. (E) C-circle assay for U2OS and SaoS2 cells after fractionation of samples into Whole, Chromatin, and Soluble fractions. DNA amount: 50 ng (F) Quantification of the C-circle assay in (E); U2OS cells as the relative amount of C-circle assay products (mean ± SD; unpaired t test), One-way ANOVA; P < 0.0001.
Furthermore, we carefully considered the temperature at which we dissolve the precipitated DNA pellet. Long incubation at high temperatures, such as 55 °C, is not optimal for the assay (SI Appendix, Fig. S1C). As previously reported, C-circles are also sensitive to temperature and freeze-thaw cycle (7). DNA dissolution temperature also affects C-circle assay due to its stability. We thus used low temperatures for DNA dissolution and avoided freeze-thaw cycle.
Single-stranded telomeres are separated via Hirt lysate, a method used for isolating extrachromosomal DNA (12, 13). Consequently, we conducted cell fractionation to investigate the presence of C-rich ssDNAs in the soluble fraction. By employing 0.1% Triton X-100, we separated the cells into a chromatin fraction (pellet) and a soluble fraction (supernatant; nucleoplasm and cytoplasm) (Fig. 1B). We validated our cell fractionation by western blot, confirming that histone H3 proteins are present in chromatin and whole fractions, GAPDH proteins are found in soluble and whole fractions, and beta-actin proteins are enriched in soluble fractions (SI Appendix, Fig. S1D).
We performed 4SET assay after cell fractionation in U2OS and SaoS2 cells, which are commonly used ALT-positive cancer cell lines derived from osteosarcoma patients (Fig. 1C and SI Appendix, Fig. S1E). As previous reports showed (12), SaoS2 cells exhibited bands around 500 bp in size that correspond to C-rich ssDNA (Fig. 1C; G-rich probe). Notably, the soluble fraction exhibited a substantial abundance of C-rich ssDNAs, while no signal was detected in the chromatin fraction, indicating that C-rich ssDNA predominantly resides in the supernatant (Fig. 1D; soluble DNA). Furthermore, our 4SET method enabled us to detect these small single-stranded C-rich telomeres in U2OS cells as well, which were not detected previously (Fig. 1C; U2OS soluble DNA). Using 30 ng DNA from soluble fractions, we were able to detect C-rich ssDNAs in U2OS cells (SI Appendix, Fig. S1F).
Extrachromosomal telomeric circular DNAs are thought to be generated in ALT-associated PML bodies (APBs). We detected that PML bodies existed in both chromatin and soluble fractions (SI Appendix, Fig. S1G). To determine whether PML bodies are responsible for the generation of C-rich ssDNA, we generated PML knockout in SaoS2 cells using CRISPR/Cas9 (SI Appendix, Fig. S1H). PML knockout SaoS2 cells do not harbor C-rich ssDNAs (SI Appendix, Fig. S1I), suggesting that C-rich ssDNAs are generated in PML bodies. Next, we measured the C-circles by using phi29 polymerase–mediated rolling circle amplification; PML knockout cells also harbor less abundant level of C-circles (SI Appendix, Fig. S1J).
We determined the size of the C-rich ssDNA using the nucleotide ladder (RiboRuler). The sizes of the C-rich ssDNAs ranged from approximately 200 to 1,500 nt (Fig. 1D). Their size distribution aligns more closely with the RiboRuler than with the base pair ladder (GeneRuler) when run on both 0.5% and 1% native agarose gels (SI Appendix, Fig. S1K), supporting that these are single-stranded DNAs.
Induction of DNA double-strand breaks using Zeocin, Etoposide (ETO), and Camptothecin (CPT) produced signals exceeding 23 kb in the soluble fraction (SI Appendix, Fig. S1L). This likely represents broken chromosomes released into the soluble fraction, supporting the idea that the soluble fraction contains extrachromosomal DNAs. Treatment with the Topoisomerase 1 inhibitor CPT, which traps the Topoisomerase 1-cleavage complex (CC), notably increased C-rich ssDNA generation, whereas ETO, targeting the Topoisomerase 2-CC, led to a milder increase. Zeocin treatment, which induces DNA double-strand breaks, did not induce this effect. This suggests that the generation of C-rich ssDNAs is more likely associated with processes such as DNA replication or transcription, rather than being a direct consequence of DNA double-strand breaks at telomeres.
Next, we measured the C-circles after cell fractionation (Fig. 1E). C-circles were also detected more in the soluble fraction (Fig. 1F and SI Appendix, Fig. S1M), which is in line with original findings on C-circles (7). We employed an alternative conventional method to fractionate cells using E1 and E2 buffers. This approach separates the cells into chromatin, nucleoplasm, and cytoplasm fractions (SI Appendix, Fig. S1 N and O). We observed that C-rich ssDNAs are enriched in both the nucleoplasm and cytoplasm fractions (SI Appendix, Fig. S1P) which aligns with the soluble fraction enrichment data (Fig. 1C).
Originating from Lagging Daughter Strands, C-Rich ssDNAs Contain Both Linear and Circular DNAs.
We conducted an S1 endonuclease digestion which specifically cleaves ssDNA. All signals in soluble fraction disappeared upon S1 nuclease treatment confirming that those are indeed ssDNA (SI Appendix, Fig. S2A). To determine whether these are linear or circular DNAs, we performed exonuclease digestion (Fig. 2A). Approximately 50% of the signals were resistant to exonucleases, suggesting that they could be circular DNAs (Fig. 2B and SI Appendix, Fig. S2B). In addition to the 4SET assay, C-rich ssDNAs in soluble fractions were confirmed as single-stranded DNA using a telomere-PNA-probe through native telomere-FISH (SI Appendix, Fig. S2C).
Fig. 2.
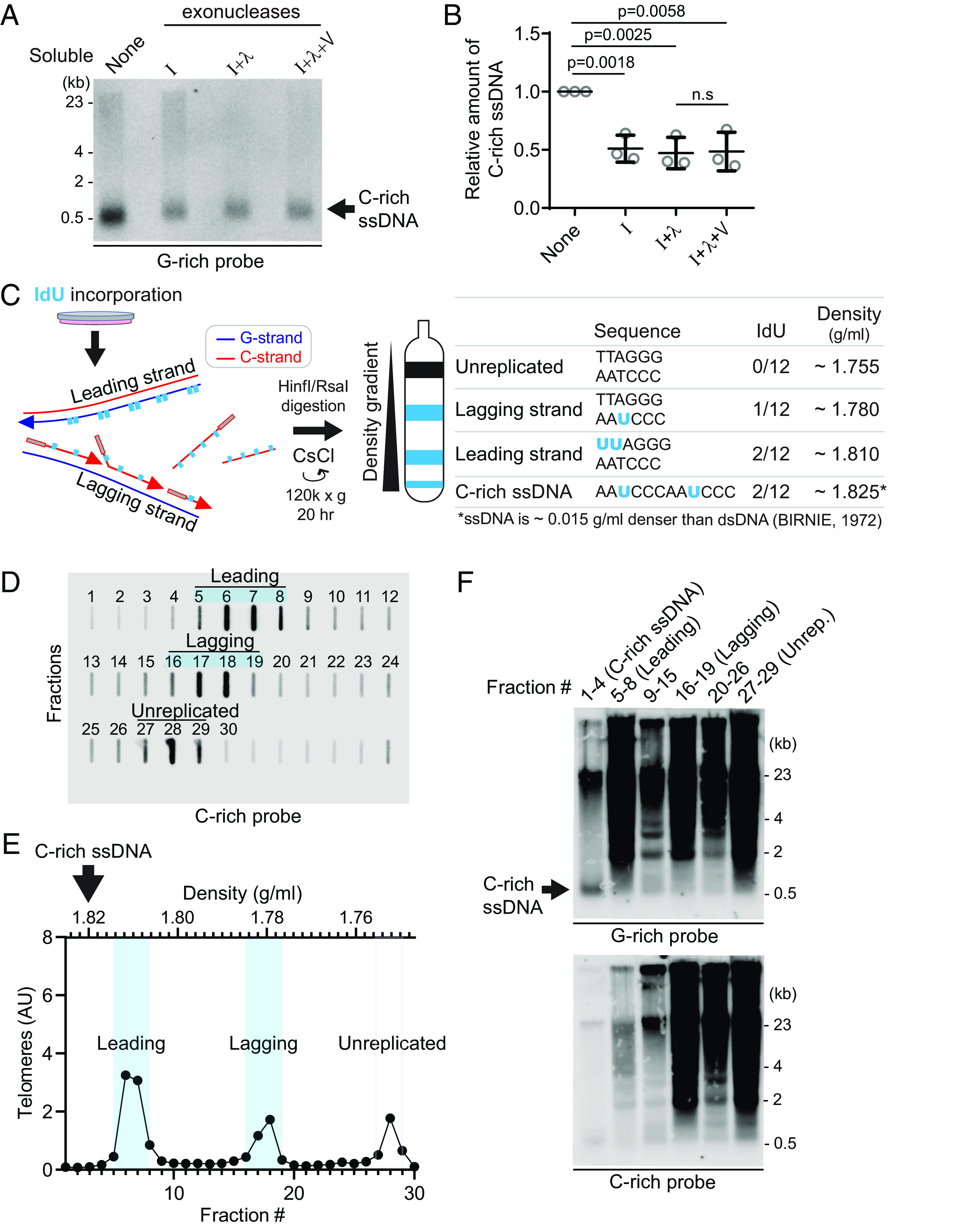
C-rich ssDNAs from lagging daughter strands contain both linear and circular DNA (A) Nuclease assay using exonuclease I, lambda, and V treatment on soluble DNA fraction of U2OS. One hundred eighty nanogram of soluble fraction DNA was used per lane. (B) Quantification of the nuclease assay in (A); Relative amount of C-rich ssDNA (mean ± SD; unpaired t test). (C) Illustration of the Iododeoxyuridine (IdU) incorporation and CsCl separation. The Right table displays the expected density of unreplicated DNA and IdU-incorporated lagging strands, leading strands, and nascent C-rich single-stranded DNAs. (D) Slot blot analysis of telomeres in CsCl separation fractions. One hundred fifty microgram of genomic DNA from U2OS cells was used. (E) Density and relative telomere intensities of fractions in (D). (F) 4SET assay of the fractions depicted in (D) and (E).
To elucidate the origin of C-rich ssDNAs, we utilized Cesium density gradient analysis (CsCl separation), a method originally used for differentiating leading and lagging strand telomeres. The incorporation of Iodo-deoxyuridine (IdU) leads to distinct density differences between these strands due to IdU’s denser nature compared to thymidine (Fig. 2 C). We hypothesized that if C-rich ssDNAs originate from lagging daughter strands, their expected density (around 1.825 g/mL) would be slightly denser than the leading strand (approximated at 1.810 g/mL) (Fig. 2C). This is because ssDNA is about 0.015 g/mL denser than dsDNA (16), despite the fact that IdU-incorporated C-rich ssDNAs and leading telomeres have same IdU-incorporated composition (2 IdU per 12 nucleotide) (Fig. 2 C; Right table). After performing CsCl separation on the genomic DNA of U2OS cells, we were able to distinguish between the leading, lagging, and unreplicated DNA fractions based on their density (Fig. 2 D and E), as we showed in our previous reports (9). Confirming our hypothesis, C-rich ssDNAs were detected around 1.825 g/mL (Fig. 2F; fraction #1 to 4), indicating that they are single-stranded and nascent DNAs possibly originating from lagging daughter strands.
MRE11 Nuclease Activity Suppresses the Generation of C-Rich ssDNA.
We investigated which nucleases are responsible for the generation of C-rich ssDNAs. We particularly focused on nucleases abundant at ALT telomeres to elucidate their role in generating C-rich ssDNAs (17). We first tested the impact of MRE11 nuclease activity inhibition in the generation of C-circles (Fig. 3A). We used Mirin, a small molecule inhibitor of MRE11 which inhibits MRE11 nuclease activity (18). Surprisingly, C-circles were increased in Mirin-treated condition in both U2OS and SaoS2 cells (Fig. 3 B and C). Next, we performed a 4SET assay in Mirin-treated condition (Fig. 3D and SI Appendix, Fig. S3A). Mirin treatment led to increases in C-rich ssDNAs mostly in the soluble fractions, with C-rich ssDNAs not detectable in the nuclear fraction (Fig. 3D; G-rich probe).
Fig. 3.
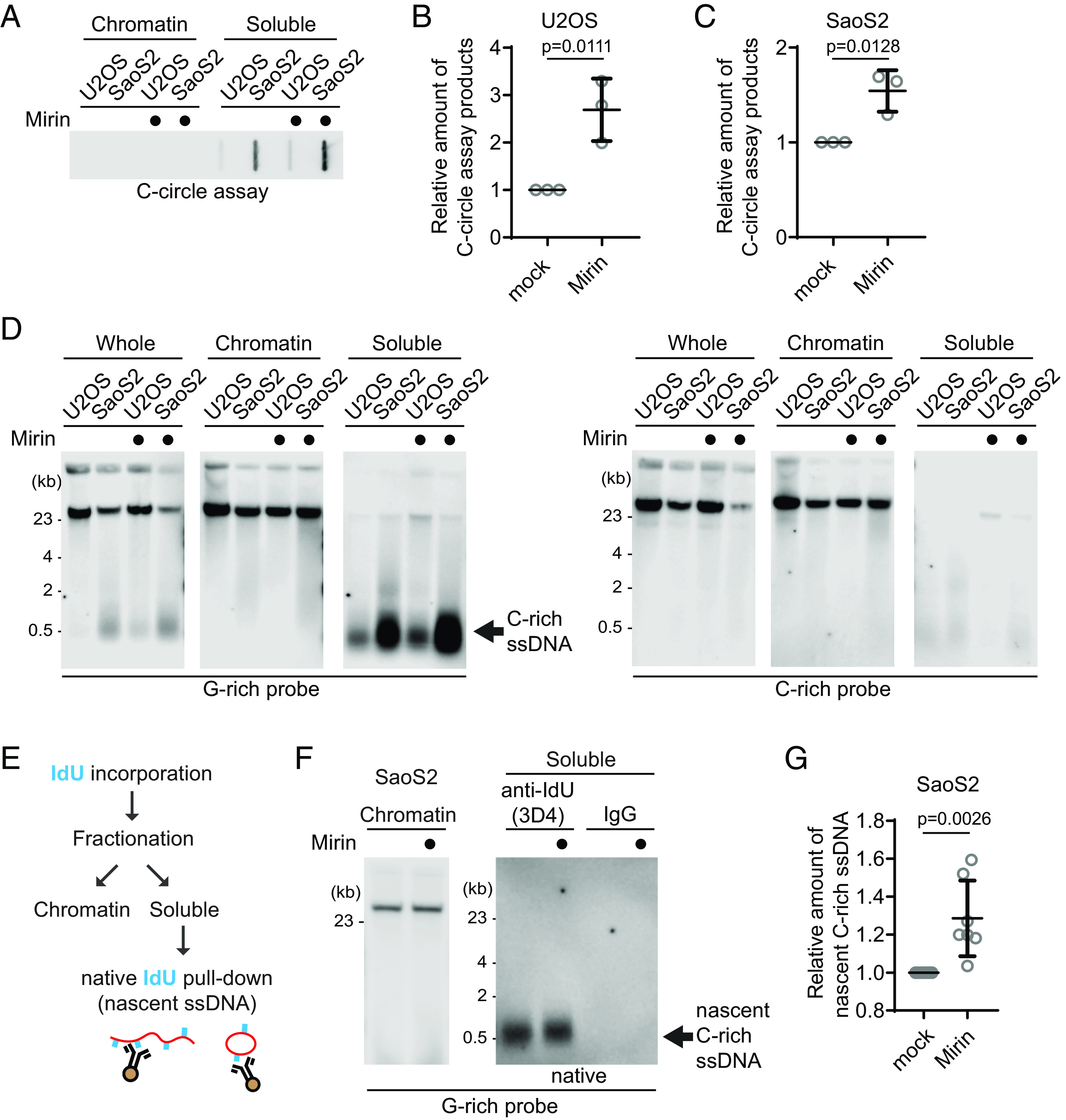
MRE11 nuclease inhibition leads to increases in C-circles and C-rich ssDNAs. (A) C-circle assay for U2OS and SaoS2 cells with or without Mirin (MRE11 nuclease inhibitor) treatment. Samples were fractionated into chromatin and soluble fractions. DNA amount: 100 ng. U2OS and Saos2 cells were treated with 50 μM Mirin for 48 h. (B and C) Quantification of the C-circle assay in A; U2OS cells (B), and SaoS2 cells (C), as the relative amount of C-circle assay products (mean ± SD; unpaired t test). (D) 4SET assay for U2OS and SaoS2 cells with or without Mirin treatment after fractionation of samples into whole, chromatin, and soluble fractions. DNA amount: 600 ng. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. (E) Illustration depicting the process of pulling down nascent single-stranded DNA from the soluble fraction incorporated with IdU. (F) Native IdU pulldown assay using IdU (3D4) antibody for SaoS2 cells treated with either Mirin (50 μM Mirin and 100 μM IdU for 20 h) or mock (100 μM IdU for 20 h). IgG-negative control; chromatin DNA-control for quantity. (G) Quantification of the native IdU-pulldown assay in (F); Relative amount of nascent C-rich ssDNAs (mean ± SD; unpaired t test).
We checked whether the impact of MRE11 inhibition in non-ALT cells to check whether Mirin-induced C-rich ssDNAs generation is ALT-specific phenotype. HeLa LT cells, telomerase-positive cells harboring long telomeres, were used (19). Mirin treatment in HeLa LT cells did not lead to generation of C-rich ssDNAs (SI Appendix, Fig. S3 B and C), indicating that C-rich ssDNAs are an ALT cancer-specific phenotype, as previously documented (12).
In addition to Mirin, we repeated the experiment for the MRE11 nuclease inhibitors PFM01 and PFM39 which are derived from Mirin structure-based chemical screening (20). Both PFM01 and PFM39 treatments also increased C-rich ssDNAs (SI Appendix, Fig. S3D), confirming that the impact of Mirin treatment is due to the inhibition of nuclease activity of MRE11. Additionally, MRE11 depletion led to a decrease in C-rich ssDNA accumulations in response to Mirin treatment (SI Appendix, Fig. S3 E–G). Introducing a mutant of MRE11 lacking nuclease activity (H129N) led to an increase in C-rich ssDNAs (SI Appendix, Fig. S3 H–J). These results further substantiate that Mirin’s impact is directly related to its inhibition of MRE11 nuclease activity.
To further validate that increased C-rich ssDNAs in Mirin treatment are nascently generated, we performed IdU pulldown in native condition of soluble DNAs to measure nascent C-rich ssDNA (Fig. 3E). Mirin treatment induces an increase in nascent C-rich ssDNA generation, indicating that MRE11 inhibition generates more C-rich ssDNAs which were nascently generated during DNA synthesis (Fig. 3 F and G).
We subsequently investigated whether PML bodies contribute to the elevated C-rich ssDNAs observed upon Mirin treatment. In PML knockout SaoS2 cells, C-rich ssDNAs were less abundant and showed no increase upon Mirin treatment (SI Appendix, Fig. S3 K and L). Moreover, these knockout cells produced significantly fewer nascent C-rich ssDNAs, indicating that the increased C-rich ssDNAs upon Mirin treatment is primarily generated at APBs (SI Appendix, Fig. S3 M and N).
C-Rich ssDNAs Are Generated during Okazaki Fragment Processing.
Prior research from both us and others has demonstrated that C-circles are produced during telomere replication and are predominantly present in the lagging strand compared to the leading strand (9, 10). Telomeres expose their G-rich ssDNAs during Okazaki fragment formation in the lagging strand (21). These G-rich ssDNAs can form higher-order structures such as G-quadruplexes, which not only contribute to telomere fragility but also serve as substrates for nucleases, such as MRE11 and DNA2 (21–23).
We observed an increase in C-rich ssDNAs in conditions where MRE11 was inhibited (Fig. 3D), possibly preventing the processing of higher-order structures in the lagging strands. Based on this observation, we hypothesized that C-rich ssDNAs are derived from lagging strand processing. To further investigate this idea, we decided to deplete DNA2, which is also involved in lagging strand processing at telomeres (23). We employed siRNA targeting DNA2 to deplete its levels in U2OS cells (SI Appendix, Fig. S4A).
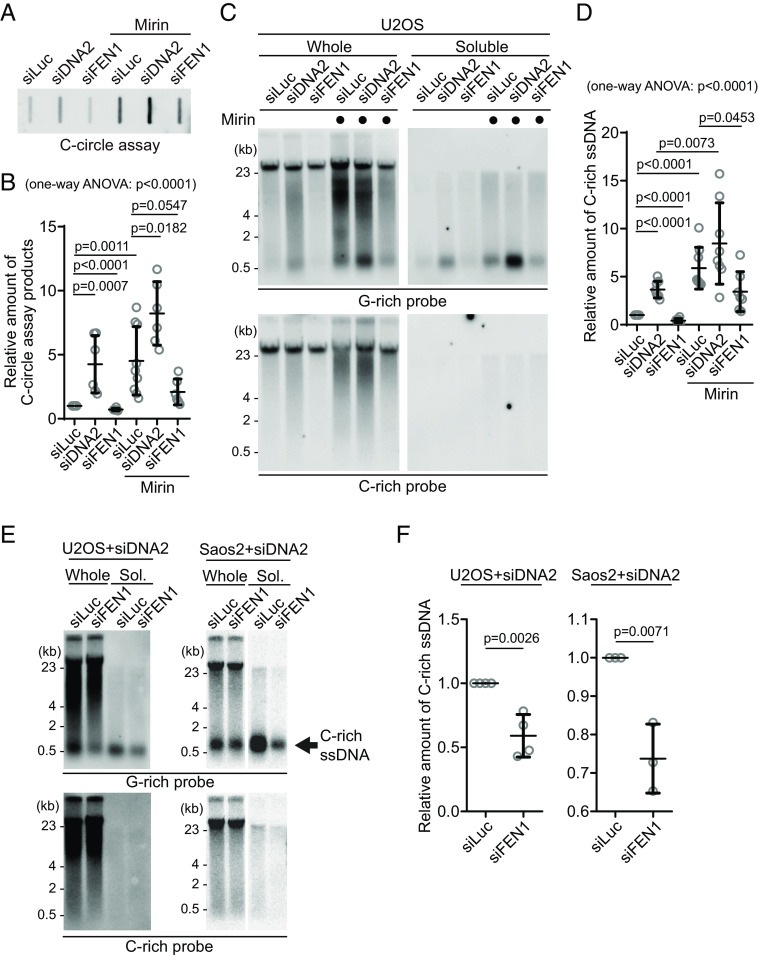
We initially measured C-circle levels in the DNA2 depletion condition (Fig. 4A) and found that, in line with previous reports, DNA2 depletion resulted in an increase in C-circle levels (Fig. 4B; siDNA2) (19). Interestingly, the combination of DNA2 depletion and Mirin treatment led to even higher C-circle levels, indicating an additive effect of inhibiting MRE11 and DNA2 depletion.
Fig. 4.
C-circles and C-rich ssDNAs are generated during Okazaki fragment processing. (A) C-circle assay for U2OS cells after transfection of siRNAs targeting DNA2 or FEN1 with or without Mirin treatment. DNA amount: 40 ng. (B) Quantification of the C-circle assay in (A) as the relative amount of C-circle assay products (mean ± SD; unpaired t test). One-way ANOVA; P < 0.0001. (C) 4SET assay for U2OS cells after transfection of siRNAs targeting FEN1, DNA2, or control (siLuc) with or without Mirin treatment. Samples were fractionated into whole DNA or soluble fraction. DNA amount (whole: 210 ng, soluble: 70 ng). G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. (D) Quantification of the assay in (C); as the relative amount of C-rich ssDNA (whole). (mean ± SD; unpaired t test). One-way ANOVA; P < 0.0001. (E) 4SET assay for U2OS and SaoS2 cells after transfection of siRNAs targeting FEN1 and DNA2, or DNA2 and siLuc. DNA amount (whole: 210 ng, soluble: 70 ng). G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. (F) Quantification of the 4SET assay in (E); as the relative amount of C-rich ssDNA (whole). (mean ± SD; unpaired t test).
Then, we measure the C-rich ssDNAs in DNA2 depleted condition. We thus conducted a 4SET assay in the DNA2 depletion condition (Fig. 4C and SI Appendix, Fig. S4B). DNA2 depletion alone caused an increase in C-rich ssDNA (Fig. 4C; siDNA2). Moreover, when Mirin was treated with DNA2 depletion, it further enhanced the accumulation of C-rich ssDNA (Fig. 4C; siDNA2 with Mirin). Therefore, in DNA2-depleted conditions, the increase in C-rich ssDNA is consistent with the increase in C-circles. However, DNA2 may not be the only nuclease involved in ssDNA generation.
As DNA2 has a role in removing the flap structures during Okazaki fragment processing, we tested whether another flap nuclease, FEN1, might be involved in C-rich ssDNA generation (24). We depleted FEN1 using siRNA (SI Appendix, Fig. S4C). Interestingly, FEN1 depletion led to a moderate decrease in C-circle levels (Fig. 4 A and B; siFEN1). In the presence of Mirin treatment, FEN1 depletion again led to partial decrease in C-circle levels (Fig. 4 A and B; siFEN1 with Mirin). Constantly, FEN1 depletion also led to a partial decrease in C-rich ssDNA generation (Fig. 4 C and D and SI Appendix, Fig. S4B) with or without Mirin treatment, which is completely opposite to our findings upon DNA2 depletion.
DNA2 exhibits a preference for cleaving long-flap structures from the 5′ end of the flap, whereas FEN1 is capable of cleaving the middle of the flap, as they are recruited by PCNA, a sliding clamp during DNA replication and repair (SI Appendix, Fig. S4D) (24). We hypothesize that FEN1 nuclease plays a role in generating C-rich ssDNAs by cutting the middle of 5′ flap structures when long-flap structures are not adequately processed by DNA2. To test this idea, we conducted a double-depletion experiment, simultaneously targeting FEN1 and DNA2, to determine whether double depletion of FEN1 and DNA2 can impede the accumulation of C-rich ssDNAs in the DNA2 depleted condition (Fig. 4 E and F and SI Appendix, Fig. S4E). Double depletion of FEN1 and DNA2 exhibited decreased C-rich ssDNAs compared to single DNA2 depletion in both U2OS and SaoS2 cells, indicating that FEN1 partially contributes to the generation of C-rich ssDNAs, possibly cleaving the flap structures in the lagging strands.
Next, we explored whether the exposed ssDNAs in lagging strand telomeres themselves could contribute to an increase in C-rich ssDNAs. To test this idea, we utilized a PARP inhibitor, which is known to impact ssDNA generation (25). We applied a modest dosage of Talazoparib, a PARP inhibitor, to induce ssDNA gaps (26). Remarkably, treatment with the PARP inhibitor resulted in an increase in C-rich ssDNAs, similar to what was observed with Mirin treatment (SI Appendix, Fig. S4F), supporting the idea that C-rich ssDNAs are generated during DNA synthesis through ssDNA exposure and gap formation.
Next, we ruled out the possibility that C-rich ssDNA’s are generated from ssDNA or gaps from the leading strand. We focused on PRIMPOL, a Primase-polymerase known for its role in repriming and gap formations in leading strands (27). Notably, depletion of PRIMPOL did not show any changes in the levels of C-rich ssDNAs (SI Appendix, Fig. S4 G and H). Furthermore, we analyzed PRIMPOL knockout cells, which have a defect in gap formation on leading strands in response to DNA replication challenges (28). When we introduced either PRIMPOL wild-type (WT) or the phosphomimic and constitutively active mutant S255D into PRIMPOL knockout cells (29) (SI Appendix, Fig. S4I), we observed no changes in C-rich ssDNAs (SI Appendix, Fig. S4J). These results also support the idea that C-rich ssDNAs are primarily derived from gap formation on the lagging strand rather than on the leading strand.
ATRX Suppresses the Generation of C-Rich ssDNAs.
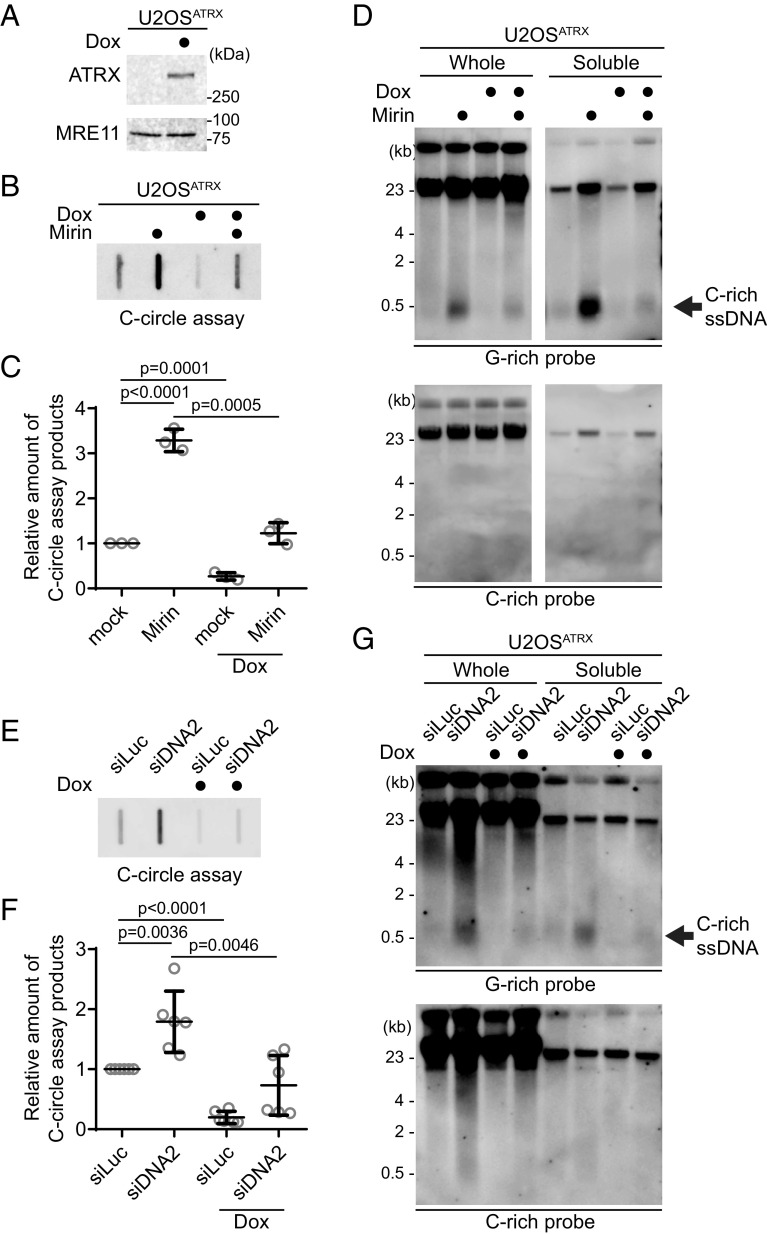
Mutations in the ATRX gene have been identified in many ALT cancers, or loss of ATRX expression is exhibited in a significant number of ALT-positive cells (30, 31). We used these U2OSATRX cells to test whether ATRX reexpression in U2OSATRX cells can affect the generation of C-rich ssDNAs (32). After 7 d of doxycycline treatment, U2OSATRX cells express ectopic ATRX (Fig. 5A).
Fig. 5.
ATRX suppresses the C-rich ssDNA generation. (A) Western blot analysis of U2OSATRX cells with ectopic ATRX expression under a doxycycline-inducible promoter, using anti-ATRX and MRE11 antibodies for the validation of ATRX expression and MRE11 levels, respectively. (B) C-circle assay for U2OSATRX cells treated with doxycycline for 1 wk or untreated, with or without Mirin treatment (50 μM Mirin for 48 h). DNA amount: 10 ng. (C) Quantification of the C-circle assay in (B) as the relative amount of C-circle assay products (mean ± SD; unpaired t test). (D) 4SET assay for U2OSATRX cells treated with doxycycline for 1 wk or untreated, with or without Mirin treatment (50 μM Mirin for 48 h). Samples were fractionated into soluble fraction or whole DNA. DNA amount (whole: 200 ng, soluble: 50 ng). G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. (E) C-circle assay for U2OSATRX cells treated with doxycycline for 1 wk or left untreated. Cells were transfected with siRNAs targeting DNA2, or control (siLuc). DNA amount: 100 ng. (F) Quantification of the C-circle assay in (B) as the relative amount of C-circle assay products (mean ± SD; unpaired t test). (G) 4SET assay for U2OSATRX cells treated with doxycycline for 1 wk or untreated. Cells were transfected with siRNAs targeting DNA2, or control (siLuc). Samples were fractionated into soluble fraction or whole DNA. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. (whole: 200 ng, soluble: 60 ng).
First, we measured C-circle levels in the doxycycline-induced ATRX expression condition (Fig. 5B). As expected, ATRX expression led to a decrease in C-circle levels. Interestingly, ATRX expression also counteracted Mirin treatment (Fig. 5 B and C; Dox with Mirin), suggesting that the increase in C-circles induced by Mirin treatment is dependent on the absence of ATRX in U2OS cells.
Subsequently, we assessed telomeric C-rich ssDNAs under the condition of ATRX expression. ATRX expression in U2OS cells decreased the quantity of C-rich ssDNAs, and it alleviated the increase of C-rich ssDNAs induced by Mirin treatment (Fig. 5D and SI Appendix, Fig. S5A). DNA2 depletion also led to an increase in C-circles in U2OSATRX cells (Fig. 5 E and F; Dox with siDNA2). However, Doxycycline-induced ATRX expression diminished the change in both C-circles and telomeric C-rich ssDNA induced by DNA2 depletion (Fig. 5G and SI Appendix, Fig. S5B). These results indicate that loss of ATRX is a crucial factor in the generation of C-rich ssDNAs in U2OS cells.
Considering that ATRX is involved in resolving G-quadruplexes (G4) at telomeres and its depletion can lead to the accumulation of R-loop structures at telomeres associated with G4 (32, 33), we predicted that DNA/RNA hybrid formation in the leading strand could produce C-rich ssDNAs by causing gaps in the unhybridized strand in the lagging strand. We investigated whether FANCM, an ATP-dependent DNA helicase/translocase involved in suppressing ALT telomere replication stress, plays a role in this process (34). We found that FANCM depletion (SI Appendix, Fig. S5C), which induces R-loop accumulation at ALT telomeres (SI Appendix, Fig. S5D) (35, 36), led to an increase in C-rich ssDNAs (SI Appendix, Fig. S5E), which is consistent with the previous observation (37). To further validate this idea, we analyzed RAD51AP1 KO cells (SI Appendix, Fig. S5F) (38), which are known to exhibit reduced G4-associated R-loop formations at ALT telomeres (39, 40). RAD51AP1 KO cells displayed lower levels of C-rich ssDNAs compared to Control cells (SI Appendix, Fig. S5G). Moreover, treatment with the G4 ligand Pyridostatin (PDS) led to an increase in C-rich ssDNAs (SI Appendix, Fig. S5 H and I). Collectively, these results indicate that loss of ATRX is a crucial factor in the generation of C-rich ssDNAs, likely facilitated by the accumulation of G4-associated R-loops at telomeres.
Nevertheless, previous studies demonstrate that the loss of ATRX genes is insufficient to induce the ALT phenotype (30), We hypothesized that C-rich ssDNA are abundant in ALT-positive cells, so we confirmed that ATRX loss alone would not generate ssDNA. As expected, when we introduced shRNA targeting the ATRX gene in HeLa LT cells, and we did not observe the generation of C-rich ssDNAs in HeLa LT cells after ATRX depletion, even when subjected to Mirin or PARPi treatment in our 24-h treatment experiment (SI Appendix, Fig. S5J). These results indicate that ATRX may not be the sole determinant in the regulation of C-rich ssDNA formation in HeLa LT cells under the specific short-term experimental conditions we employed.
CST Complex-Mediated Priming and Subsequent Strand Displacements by DNA Polymerase Delta Generate C-Rich ssDNAs.
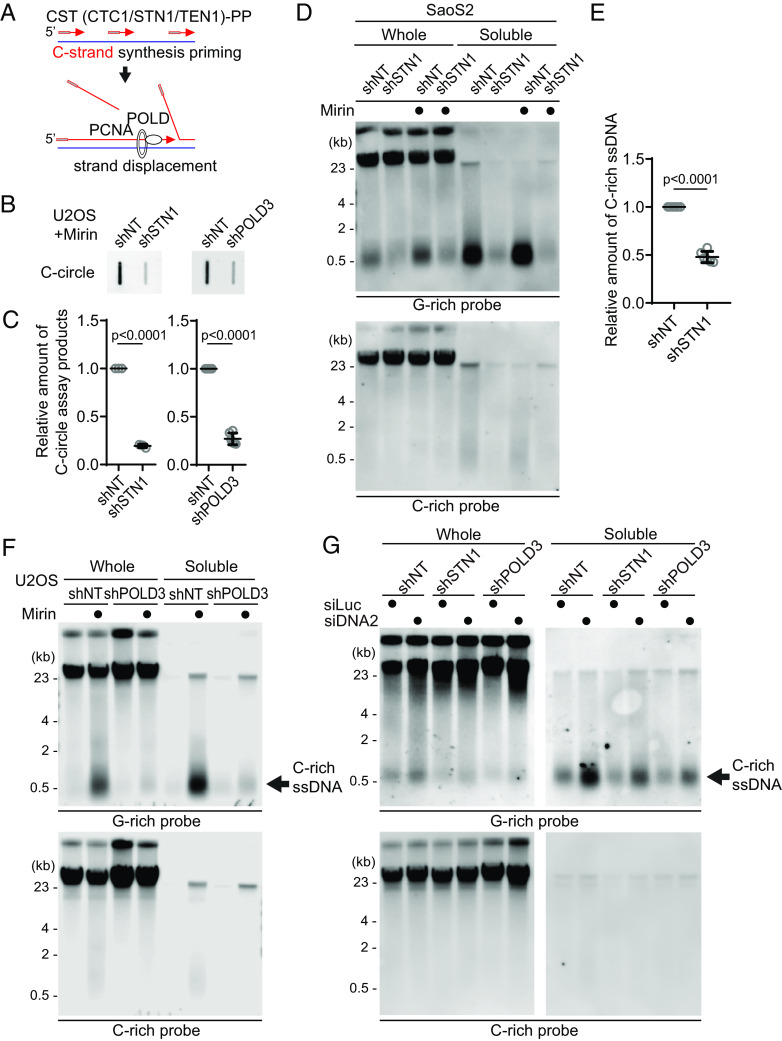
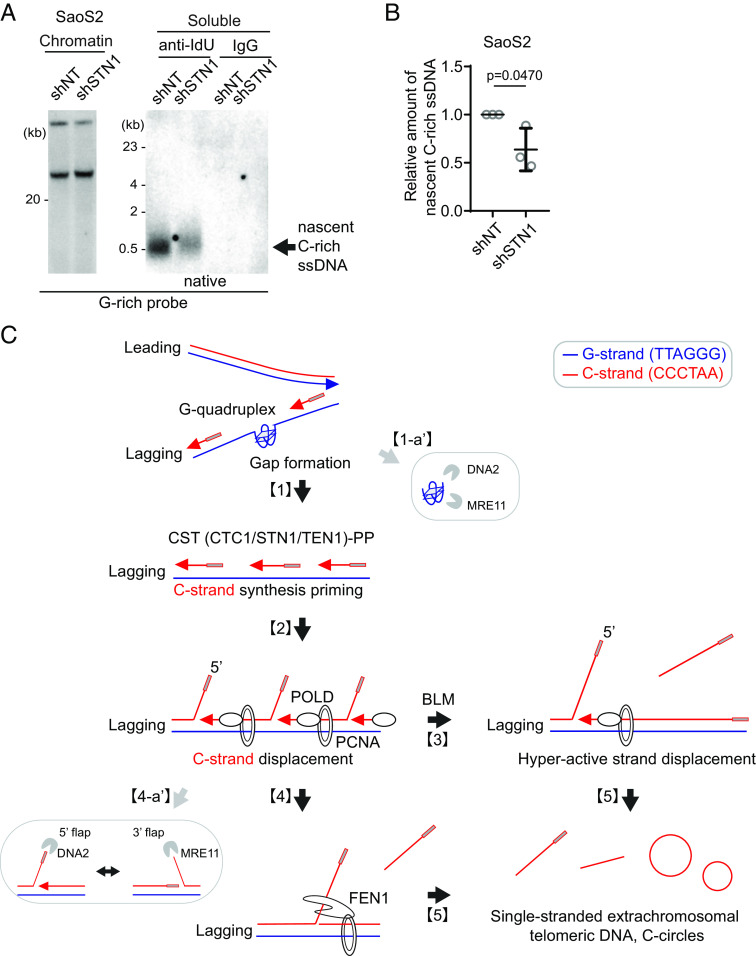
The CST (CTC1/STN1/TEN1)-PP (Primase/DNA polymerase alpha) complex plays a crucial role in the C-strand fill-in processes at telomeres (41). It recognizes exposed G-rich strands, including higher-order structures such as G-quadruplex, on the parental lagging strand during telomere replication and initiates the filling in of the complementary C-strand, generating additional Okazaki fragments that serve to remove the gaps in the lagging strand and ensure the telomere replication (42, 43). Because the exposed ssDNAs in lagging strand telomeres are priming sites for CST complex, we hypothesized that C-rich ssDNAs could be generated by additional Okazaki fragments formation by CST complex-mediated DNA priming (Fig. 6A). We depleted STN1, a component of CST complex, using shRNA targeting STN1 (43) (SI Appendix, Fig. S6A). In line with the previous report, depletion of STN1 led to a reduction of C-circles in Mirin-treated U2OS cells (Fig. 6 B and C). We performed a 4SET assay to measure the levels of C-rich ssDNAs in conditions where STN1 was depleted. Surprisingly, the depletion of STN1 led to a striking reduction in C-rich ssDNAs in both U2OS and SaOS2 cells (Fig. 6 D and E and SI Appendix, Fig. S6 B and C). Furthermore, STN1 depletion also resulted in the dramatic abolishment of C-rich ssDNAs in response to Mirin treatment. These findings suggest that STN1 plays a crucial role in the generation of C-rich ssDNAs and highlights its involvement in the response to Mirin treatment in these cells.
Fig. 6.
C-circles and C-rich ssDNAs are generated through C-strand priming and subsequent strand displacement. (A) Illustration depicting C-strand synthesis priming by CST-PP (CTC1/STN1/TEN1-Primase-Polymerase alpha)-complex and the strand displacement of Okazaki fragments by PCNA and DNA polymerase delta (POLD) during DNA replication on the lagging strand. (B) C-circle assay for U2OS cells expressing shRNAs targeting STN1, POLD3, or nontargeting (NT) control. Cells were treated with Mirin (50 μM, 48 h) DNA amount: Left—100 ng, Right—30 ng. (C) Quantification of the C-circle assay in (B) as the relative amount of C-circle assay products (mean ± SD; unpaired t test). (D) 4SET assay for SaoS2 expressing shRNAs targeting STN1, or nontargeting (NT) control. Cells were treated with Mirin (50 μM, 48 h). Samples were fractionated into soluble fraction or whole DNA. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. DNA amount (Saos2 whole: 200 ng, soluble: 60 ng). (E) Quantification of the 4SET assay in (D); as the relative amount of C-rich ssDNA (whole). (mean ± SD; unpaired t test). (F) 4SET assay for U2OS cells expressing shRNAs targeting POLD3, or nontargeting (NT) control. Samples were fractionated into soluble fraction or whole DNA. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. DNA amount (whole: 300 ng, soluble: 100 ng). (G) 4SET assay for U2OS cells expressing shRNAs targeting STN1, POLD3, or nontargeting (NT) control after transfection of siRNAs targeting DNA2 or control (siLuc). Samples were fractionated into soluble fraction or whole DNA. G-rich probe was used to detect C-rich telomeric sequences, and C-rich probe was used to detect G-rich telomeric sequences. C-rich single-stranded DNAs are indicated by an arrow. DNA amount (whole: 210 ng, soluble: 70 ng).
Primed Okazaki fragments, created by DNA polymerase alpha, are subsequently extended by DNA polymerase delta (24). This sequential process ensures the continuous synthesis of the lagging strand during DNA replication and creates a flap structure as polymerase delta displaces the downstream DNA strand. Particularly in ALT cells, the excessive strand displacement activity of polymerase delta, working with the BLM helicase and other associated factors, plays a crucial role in ALT telomere elongation through BIR (4, 44–46). The helicase activity of BLM is crucial for these processes (44). We hypothesized that the primed C-strand Okazaki fragments, initiated by the CST-PP complex, undergo prolonged extension by DNA polymerase delta and BLM helicase, creating a flap structure. This unique excessive strand displacement activity in ALT cells allows for the complete displacement of downstream DNA, resulting in the accumulation of C-rich ssDNA (Fig. 6A).
To test this idea, we depleted POLD3, a component of DNA polymerase delta, using shRNA targeting POLD3 (47, 48) (SI Appendix, Fig. S6D). Depletion of POLD3 led to a decrease in C-circles in Mirin-treated U2OS cells. We conducted a 4SET assay to measure the C-rich ssDNAs in POLD3-depleted cells. Strikingly, POLD3-depletion reduced the C-rich ssDNAs in U2OS cells (Fig. 6F and SI Appendix, Fig. S6E). Moreover, POLD3-depletion also abolished the C-rich ssDNA in response to Mirin treatment.
To further validate the involvement of DNA polymerase alpha and delta in the generation of C-rich ssDNAs, we conducted 4SET assay after treating low dosages of Aphidicolin (a DNA polymerase alpha/delta inhibitor) and CD437 (a DNA polymerase alpha inhibitor) to partially inhibit their actions. The results were in line with our hypothesis, as both Aphidicolin and CD437 treatments led to decreases in C-rich ssDNAs (SI Appendix, Fig. S6F), confirming that C-rich ssDNAs are indeed derived from lagging strand processes mediated by DNA polymerase alpha and delta. The inhibition of these DNA polymerases resulted in reduced C-rich ssDNA formation, supporting their role in this process during telomere replication.
As we hypothesized, we observed that BLM knockout cells showed reduced levels of C-rich ssDNAs (SI Appendix, Fig. S6 G and H). These results support that BLM helicase is involved in the generation of C-rich ssDNAs in the context of ALT telomere replication through BIR. The reduced levels of C-rich ssDNAs upon BLM depletion suggest that BLM’s excessive strand displacement activity likely plays a crucial role in this process.
Furthermore, STN1- and POLD3-depletion also mitigated the increase of C-rich ssDNAs induced by DNA2 depletion (Fig. 6G and SI Appendix, Fig. S6I), supporting the idea that C-strand synthesis priming followed by excessive strand displacement is required for the generation of C-rich ssDNAs in ALT cells. Finally, we assessed nascent C-rich ssDNA under STN1-depleted conditions (Fig. 7A). The depletion of STN1 resulted in reduced nascent C-rich ssDNA (Fig. 7B), again demonstrating that C-rich ssDNAs are nascently generated and arise from CST-complex mediated priming at the lagging strands.
Fig. 7.
Origins of C-circles and their precursor linear C-rich single-stranded DNAs: Excessive strand displacements from lagging strand telomeres. (A) Native IdU pulldown was conducted using IdU (3D4) antibody for SaoS2 cells expressing shRNAs targeting STN1, or nontargeting (NT) control. IgG served as a negative control. Chromatin DNA was also included to control for DNA quantity. (B) Quantification of the native IdU-pulldown assay in (A); Relative amount of nascent C-rich ssDNAs (mean ± SD; unpaired t test). (C) Working model: [1] Accumulations of G-quadruplex (G4) structures and G4-associated R-loops at telomeres may contribute to the generation of C-rich single-stranded DNAs (ssDNAs) by potentially exposing parental lagging strands, which can be recognized by the CST-PP (CTC1-STN1-TEN1-Polα-Primase) complex. [2] The CST-PP complex exhibits priming activity in the exposed gaps or G4 structures in lagging strands, which are subsequently extended by DNA polymerase delta. [3] As a result, primed C-strand Okazaki fragments are initiated by the CST-PP complex and undergo prolonged extension facilitated by DNA polymerase delta and BLM helicase, leading to the formation of long flap structures. [4] In the context where long flap structures are not adequately processed by DNA2, the FEN1 nuclease may contribute to the generation of C-rich ssDNAs by cleaving the middle of 5′ flap structures. [5] In parallel, the unique excessive strand displacement activity observed in ALT cells enables the complete displacement of downstream DNA, thereby promoting the accumulation of C-rich single-stranded DNA. DNA2 and MRE11 are potentially involved in lagging strand telomere processes by participating in the removal of G4 structures [1-a′] and processing flap structures in Okazaki fragments on the lagging strands [4-a′].
Discussion
Here, our study demonstrated that inhibiting the nuclease activity of MRE11 results in an increase in the generation of C-rich ssDNAs and C-circles. These findings suggest that the nuclease activity of MRE11 plays a role in regulating the levels of C-rich ssDNAs and C-circles, potentially contributing to the resolution of higher-order structures, such as G-quadruplexes in lagging strands (Fig. 7C; [1-a’]) or 5’ flap structures in Okazaki fragments (Fig. 7C; [4-a’]). Notably, yeast genetics showed that mre11 and the mre11 nuclease mutant are synthetic lethal with rad27 (yeast homolog of human FEN1) (49), even though in vitro biochemistry experiments did not reveal preferential MRE11 nuclease activity for flap structures (50).
Additionally, our study demonstrated that depletion of DNA2 leads to an increase in the generation of C-rich ssDNAs, consistent with previous reports that showed elevated C-circles and APBs upon DNA2 depletion (19). DNA2 plays a crucial role in resolving G-quadruplex structures in lagging strand telomeres, which likely contributes to the observed accumulation of C-rich ssDNAs upon DNA2 depletion (Fig. 7C; [1-a’]).
DNA2 also plays a crucial role in Okazaki fragment processing, particularly in the removal of flap structures (24). Recent structural studies have unveiled that DNA2 adopts a cylindrical shape with a tunnel through which ssDNA passes, and its nuclease domain is embedded within this tunnel (51). DNA2 efficiently trims ssDNA from the ssDNA end and preferentially processes long flaps (Fig. 7C; [4-a’]). This process relies on its interaction with RPA (Replication Protein A) (51). We propose that DNA2 depletion results in an increased presence of long flaps, which are partly processed by FEN1 or engaged in strand displacement by DNA polymerase delta. FEN1 nuclease may partly contribute to the generation of C-rich ssDNAs by cleaving the middle of 5′ flap structures when long-flap structures are not adequately processed by DNA2 (Fig. 7C; [4]). However, the contribution of FEN1-mediated cleavage in C-rich ssDNAs may not be substantial when the flaps are long and bound by RPA. Indeed, further study is essential to fully elucidate the precise mechanisms by which these enzymes coordinate their activities for Okazaki fragment processes in the lagging strand, particularly at ALT telomeres.
Our results indicate that the loss of ATRX is a crucial factor in the generation of C-rich ssDNAs, likely facilitated by the accumulations of G-quadruplex (G4) structures and G4-associated R-loops at telomeres. This is supported by FANCM depletion and RAD51AP1 KO data showing that R-loop formation may contribute to the generation of C-rich ssDNAs, possibly by exposing parental lagging strands that can be recognized by the CST-PP complex (Fig. 7C; [1]).
The CST-PP complex has critical roles during telomere replication in particular of the lagging strand (43). Previous reports demonstrated that CST complex has a role in resolving G4 structures at telomeres as well as genome-wide (52). Interestingly, CST complex is detected in APBs (42). Depletion of CST leads to replication defects in ALT cells, accompanied by decreases in T-circles and C-circles (42). We interpret that CST’s priming activity in exposed gap or G4 structures in lagging strands is responsible for the generation of C-rich ssDNAs and C-circles. These primed Okazaki fragments, created by DNA polymerase alpha, are subsequently extended by DNA polymerase delta (Fig. 7C; [2]).
In ALT cells, the interplay between polymerase delta’s excessive strand displacement activity, along with BLM helicase and other associated factors, plays a crucial role in ALT telomere elongation through Break-Induced Replication. We propose that primed C-strand Okazaki fragments, initiated by the CST-PP (CTC1-STN1-TEN1-Polα-Primase) complex, undergo prolonged extension facilitated by the DNA polymerase delta and the BLM helicase, resulting in the formation of long flap structures (Fig. 7C; [3]). The unique excessive strand displacement activity in ALT cells enables the complete displacement of downstream DNA, leading to the accumulation of C-rich single-stranded DNA (Fig. 7C; [5]).
Materials and Methods
Cell lines in all experiments were authenticated prior to use. Experiments have been conducted at least three times, and biological inferences were drawn only when independent trials produce the same outcome. The experiments encompass a series of procedures, cell culture, siRNA/shRNA-mediated knockdown, and sgRNA knockout. These are complemented by plasmid construction and utilization, 4SET Assay, and CsCl separation for telomeric strand analysis. Further, we employed cell fractionation, IdU Pull-down for nascent C-rich ssDNA isolation, and the C-circle assay for detection of C-circles. This study also involved the use of specific antibodies, RNA extraction and quantification for gene expression analysis, and statistical analyses for validation. Detailed protocols for the experiments are extensively documented and available in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported by the following NIH/NCI grants: CA207209 (R.J.O.), CA262316 (R.J.O.), CA197774 (A.C.), and CA245259 (J.M.).
Author contributions
J.M. designed research; Junyeop Lee, Jina Lee, E.J.S., and J.M. performed research; A.T., R.J.O., A.C., and J.M. contributed new reagents/analytic tools; J.M. analyzed data; and J.M. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Bryan T. M., Englezou A., Dalla-Pozza L., Dunham M. A., Reddel R. R., Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Lydeard J. R., Jain S., Yamaguchi M., Haber J. E., Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448, 820–823 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Roumelioti F. M., et al. , Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 17, 1731–1737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dilley R. L., et al. , Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 539, 54–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Min J., Wright W. E., Shay J. W., Alternative lengthening of telomeres mediated by mitotic DNA synthesis engages break-induced replication processes. Mol. Cell Biol. 37, e00226-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesare A. J., Reddel R. R., Alternative lengthening of telomeres: Models, mechanisms and implications. Nat. Rev. Genet. 11, 319–330 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Henson J. D., et al. , DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat. Biotechnol. 27, 1181–1185 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Henson J. D., et al. , The C-circle assay for alternative-lengthening-of-telomeres activity. Methods 114, 74–84 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Min J., Wright W. E., Shay J. W., Alternative lengthening of telomeres can be maintained by preferential elongation of lagging strands. Nucleic Acids Res. 45, 2615–2628 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang T., et al. , Strand break-induced replication fork collapse leads to C-circles, C-overhangs and telomeric recombination. PLoS Genet. 15, e1007925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu R., Pickett H. A., Telomeric replication stress: The beginning and the end for alternative lengthening of telomeres cancers. Open Biol. 12, 220011 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nabetani A., Ishikawa F., Unusual telomeric DNAs in human telomerase-negative immortalized cells. Mol. Cell Biol. 29, 703–713 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oganesian L., Karlseder J., Mammalian 5’ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol. Cell 42, 224–236 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oganesian L., Karlseder J., 5’ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst) 12, 238–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai T. P., Wright W. E., Shay J. W., Generation of digoxigenin-incorporated probes to enhance DNA detection sensitivity. Biotechniques 60, 306–309 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Birnie G. D., Separation of native and denatured DNA, RNA and hybrid on sodium iodide gradients. FEBS Lett. 27, 19–22 (1972). [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Exposito L., et al. , Proteomic profiling reveals a specific role for translesion DNA polymerase eta in the alternative lengthening of telomeres. Cell Rep. 17, 1858–1871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupre A., et al. , A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4, 119–125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Sullivan R. J., et al. , Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nat. Struct. Mol. Biol. 21, 167–174 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shibata A., et al. , DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol. Cell 53, 7–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnoult N., Saintome C., Ourliac-Garnier I., Riou J. F., Londono-Vallejo A., Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 23, 2915–2924 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J., et al. , Dynamic interaction of BRCA2 with telomeric G-quadruplexes underlies telomere replication homeostasis. Nat. Commun. 13, 3396 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W., et al. , Mammalian DNA2 helicase/nuclease cleaves G-quadruplex DNA and is required for telomere integrity. EMBO J. 32, 1425–1439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun H., et al. , Okazaki fragment maturation: DNA flap dynamics for cell proliferation and survival. Trends Cell Biol. 33, 221–234 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cong K., et al. , Replication gaps are a key determinant of PARP inhibitor synthetic lethality with BRCA deficiency. Mol. Cell 81, 3128–3144.e3127 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Y., et al. , BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 19, 5003–5015 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taglialatela A., et al. , REV1-Polzeta maintains the viability of homologous recombination-deficient cancer cells through mutagenic repair of PRIMPOL-dependent ssDNA gaps. Mol. Cell 81, 4008–4025.e4007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinet A., et al. , PRIMPOL-mediated adaptive response suppresses replication fork reversal in BRCA-deficient cells. Mol. Cell 77, 461–474.e469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta K. P. M., et al. , CHK1 phosphorylates PRIMPOL to promote replication stress tolerance. Sci. Adv. 8, eabm0314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lovejoy C. A., et al. , Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8, e1002772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heaphy C. M., et al. , Altered telomeres in tumors with ATRX and DAXX mutations. Science 333, 425 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clynes D., et al. , Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 6, 7538 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teng Y. C., et al. , ATRX promotes heterochromatin formation to protect cells from G-quadruplex DNA-mediated stress. Nat. Commun. 12, 3887 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X., et al. , FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc. Natl. Acad. Sci. U.S.A. 114, E5940–E5949 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan X., et al. , FANCM suppresses DNA replication stress at ALT telomeres by disrupting TERRA R-loops. Sci. Rep. 9, 19110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva B., et al. , FANCM limits ALT activity by restricting telomeric replication stress induced by deregulated BLM and R-loops. Nat. Commun. 10, 2253 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu R., et al. , The FANCM-BLM-TOP3A-RMI complex suppresses alternative lengthening of telomeres (ALT). Nat. Commun. 10, 2252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barroso-Gonzalez J., et al. , RAD51AP1 is an essential mediator of alternative lengthening of telomeres. Mol. Cell 76, 11–26.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yadav T., et al. , TERRA and RAD51AP1 promote alternative lengthening of telomeres through an R- to D-loop switch. Mol. Cell 82, 3985–4000.e3984 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaminski N., et al. , RAD51AP1 regulates ALT-HDR through chromatin-directed homeostasis of TERRA. Mol. Cell 82, 4001–4017.e4007 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai S. W., de Lange T., CST-Polalpha/Primase: The second telomere maintenance machine. Genes Dev. 37, 555–569 (2023), 10.1101/gad.350479.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C., Jia P., Chastain M., Shiva O., Chai W., The human CTC1/STN1/TEN1 complex regulates telomere maintenance in ALT cancer cells. Exp. Cell Res. 355, 95–104 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang C., Dai X., Chai W., Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22, 1681–1695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Min J., Wright W. E., Shay J. W., Clustered telomeres in phase-separated nuclear condensates engage mitotic DNA synthesis through BLM and RAD52. Genes Dev. 33, 814–827 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobinoff A. P., et al. , BLM and SLX4 play opposing roles in recombination-dependent replication at human telomeres. EMBO J. 36, 2907–2919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loe T. K., et al. , Telomere length heterogeneity in ALT cells is maintained by PML-dependent localization of the BTR complex to telomeres. Genes Dev. 34, 650–662 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li S., et al. , PIF1 helicase promotes break-induced replication in mammalian cells. EMBO J. 40, e104509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min J., et al. , Mechanisms of insertions at a DNA double-strand break. Mol. Cell 83, 2434–2448.e7 (2023), 10.1016/j.molcel.2023.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreau S., Ferguson J. R., Symington L. S., The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol. Cell Biol. 19, 556–566 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paull T. T., Gellert M., The 3’ to 5’ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol. Cell 1, 969–979 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Zhou C., Pourmal S., Pavletich N. P., Dna2 nuclease-helicase structure, mechanism and regulation by Rpa. Elife 4, e09832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li T., et al. , Cooperative interaction of CST and RECQ4 resolves G-quadruplexes and maintains telomere stability. EMBO Rep. 24, e55494 (2023), 10.15252/embr.202255494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.