Abstract
Pericentromeric heterochromatin mostly comprises repeated DNA sequences prone to ectopic recombination. In Drosophila cells, ‘safe’ homologous recombination repair requires relocalization of heterochromatic repair sites to the nuclear periphery before Rad51 recruitment and strand invasion. DSBs are anchored to the nuclear periphery through the Nup107/160 nucleoporin complex. Previous studies suggested that the nuclear pore ‘basket’ protein Nup153 could also mediate anchoring, but Nup153 RNAi depletion also affects Nup107 association with the pores, preventing a direct assessment of Nup153 role. Using a separation of function mutant, here we show that Nup153 is not required for anchoring heterochromatic DSBs to the nuclear periphery.
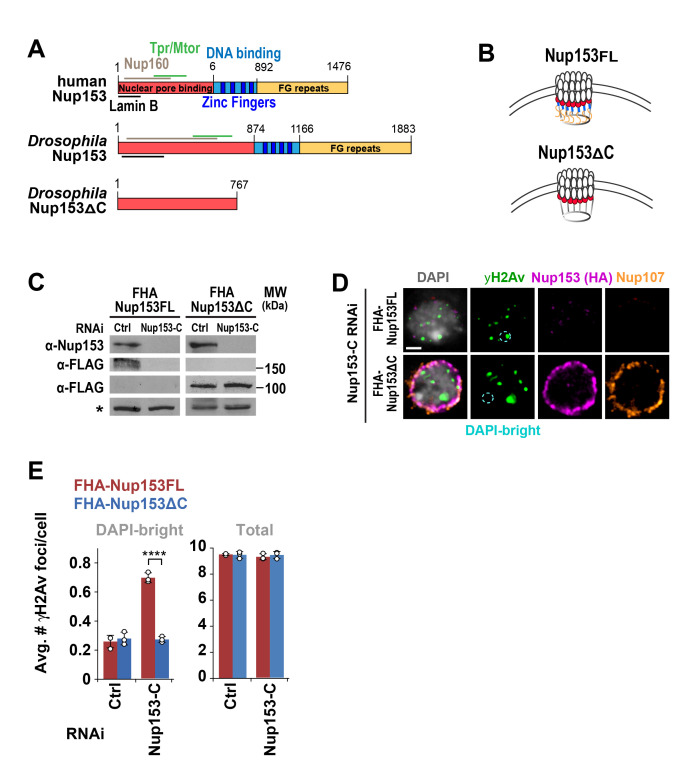
Figure 1. Nup153 DNA-binding and FG-repeat domains are not required for relocalization of heterochromatic DSBs.
A: Scheme of human and Drosophila Nup153 protein, highlighting the domains mapped in the human protein and conserved domains in the Drosophila protein. The N-terminus is responsible for binding other pore proteins and Lamina B, as indicated. B: Schematic representation of Nup153 FL and ΔC at the pore. C: Western blot analysis with the indicated antibodies of cells expressing FLAG-HA(FHA)-Nup153FL and FHA-Nup153ΔC shows efficient depletion of endogenous Nup153 and FHA-Nup153FL. (*) A non-specific band is used as a loading control. D: Immunofluorescence (IF) of indicated proteins in cells expressing FHA-Nup153FL or FHA-Nup153ΔC after RNAi depletion of endogenous Nup153 and FHA-Nup153FL. Scale bar = 1µm. E: Quantification of γH2Av foci in DAPI-bright and total foci in cells expressing FHA-Nup153FL or FHA-Nup153ΔC fixed 60 min after IR, following indicated RNAi depletions. Nup153-C indicates that siRNAs recognize the C-terminal region of the protein. ****P < 0.0001, two-tailed Mann-Whitney test, n > 166 cells from three independent experiments. Error bars: mean +/- SD.
Description
Pericentromeric heterochromatin (hereafter "heterochromatin") constitutes about 30% and 10% of fly and human genomes respectively, and is mostly composed of repeated DNA sequences (Lander et al. 2001, Hoskins et al. 2007, Ho et al. 2014, Hoskins et al. 2015, Nurk et al. 2022) . In heterochromatin, thousands to millions of identical DNA repeats, including from different chromosomes, can engage in ectopic recombination (Peng and Karpen 2008, Amaral et al. 2017) , presenting a serious threat to genome stability in multicellular eukaryotes. In Drosophila , heterochromatin forms a distinct nuclear domain (Chiolo et al. 2011, Riddle et al. 2011, Li et al. 2017, Caridi et al. 2018, See et al. 2021) and aberrant recombination is prevented by relocalization of double-strand breaks (DSBs) to the nuclear periphery before Rad51 recruitment and strand invasion (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016, Caridi et al. 2018, Merigliano et al. 2023, Miller et al. 2023) . Early HR steps occur within the heterochromatin domain, while later steps are initially blocked and resume only after relocalization (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016, Caridi et al. 2018, Merigliano et al. 2023) .
This movement relies on the SUMO E3 ligase Smc5/6 complex (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016) and its interactors: the actin nucleator Arp2/3 (Caridi et al. 2018) and nuclear myosins (Caridi et al. 2018) . Arp2/3 coordinates the formation of nuclear actin filaments that start polymerizing at heterochromatic DSBs and enable the directed, myosin-driven, movement of repair sites to the nuclear periphery (Caridi et al. 2018) . The mobilization of heterochromatic repair sites also requires nucleoporins associated with the chromatin in the nucleoplasm, particularly the phase separation properties of Nup98 (Merigliano et al. 2023) . Nup98 creates a condensate inside the heterochromatin domain, facilitating the initial diffusive movement of repair sites from the core to the periphery of the heterochromatin domain, where capturing from nuclear F-actin occurs (Merigliano et al. 2023) . Once at the nuclear periphery, repair sites associate with the nuclear pores through their interaction with the Y complex, including Nup107 and Nup160 (Ryu et al. 2015) . These might enable repair restart through the degradation of SUMOylated targets mediated by the SUMO-targeted ubiquitin ligase (STUbL) Dgrn (Ryu et al. 2015) .
Relocalization likely prevents aberrant recombination by separating damaged DNAs from ectopic repeated sequences, thus promoting ‘safe’ exchanges with the sister chromatid or the homologous chromosome (Caridi et al. 2017, Caridi et al. 2019, Rawal et al. 2019) . Accordingly, loss of components required for relocalization results in persistent unrepaired heterochromatic DSBs and widespread chromosome rearrangements (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016, Caridi et al. 2018, Dialynas et al. 2019, Merigliano et al. 2023) . Similar mechanisms have been described in mammalian cells (Jakob et al. 2011, Chiolo et al. 2013, Tsouroula et al. 2016, Caridi et al. 2018) , revealing conserved pathways.
In Drosophila cells, repair sites leave the heterochromatin domain 10 min after DSB induction with ionizing radiation (IR). This results in fewer repair sites (γH2Av foci) in DAPI-bright heterochromatin (a subsection of the heterochromatin domain (Chiolo et al. 2011) ) 60 min after IR (Chiolo et al. 2011, Ryu et al. 2015, Caridi et al. 2018, Merigliano et al. 2023) . In the absence of Nup107, defective anchoring to the nuclear periphery results in a higher number of γH2Av foci in DAPI-bright 60 min after IR, without affecting total focus count (Ryu et al. 2015) . A similar result is observed after RNAi depletion of the nuclear pore basket protein Nup153 (Ryu et al. 2015) . However, Nup153 RNAi also results in significant loss of the nuclear periphery signal of Nup107 (Ryu et al. 2015) , consistent with a role for Nup153 in nuclear pore assembly and Nup107 recruitment (Vollmer et al. 2015) . Thus, the effect of Nup153 RNAi on relocalization of heterochromatic DSBs could be indirect, through the loss of Nup107 (Ryu et al. 2015) .
Here we assessed the role of Nup153 in heterochromatin repair by expressing in Drosophila cells a Nup153∆C mutant, which retains the N-terminal domain required for Nup107 recruitment while losing DNA binding and pore transport domains (Griffis et al. 2004) ( Figure 1A,B ). As a control, we generated cells expressing Nup153 full-length (FL). We RNAi depleted Nup153 by targeting its C-terminal region (Nup153-C RNAi), which affects the levels of endogenous Nup153 and FHA-Nup153FL, without altering FHA-Nup153∆C ( Figure 1C ). As expected, RNAi depletion of Nup153 results in significant loss of Nup107 signal at the nuclear periphery ( Figure 1D ) (Ryu et al. 2015) . However, cells display normal nuclear size and decondensed chromatin ( Figure 1D ), suggesting that other components of the pore are intact (Boehmer et al. 2003) or that residual Nup107 is present to sustain some pore assembly (Vollmer et al. 2015) . Importantly, expression of Nup153∆C in cells depleted for endogenous Nup153 fully restores Nup107 signal at the pores ( Figure 1D ), consistent with the Y complex being intact.
Next, we tested how the loss of most Nup153 domains affects heterochromatin repair by investigating the distribution of repair foci relative to DAPI-bright in cells expressing Nup153∆C. RNAi depletion of endogenous Nup153 and FHA-Nup153FL results in a higher number of γH2Av foci in DAPI-bright heterochromatin 60 min after IR, consistent with a relocalization defect and previous studies (Ryu et al. 2015) . However, expression of FHA-Nup153∆C in the absence of endogenous Nup153 lowers the number of γH2Av foci in DAPI-bright 60 min after IR to a level similar to that observed in control RNAi cells, without affecting total focus count ( Figure 1D ). This is consistent with normal relocalization of heterochromatic DSBs. Thus, focus relocalization occurs normally in cells lacking all the N-terminal Nup153 functional domains, including DNA-binding activities and FG repeats. We conclude that Nup153 is not required for relocalizing heterochromatic DSBs, and thus it does not contribute to anchoring heterochromatic repair sites to the nuclear periphery.
Methods
Cell cultures
Kc167 (Kc) cells were used for all experiments and were maintained as logarithmically growing cultures in Schneider’s medium (Sigma) or Sf-900 II (ThermoFisher) + 10% FBS (GemCell US Origin, Gemini) + antibiotic-antimycotic (ThermoFisher). Kc cells were authenticated by the Drosophila Genomic Resource Center (DGRC) and no mycoplasma contamination was detected.
Generation of cell lines expressing tagged proteins
Experiments were performed using stable cell lines obtained by cotransfecting each plasmid of interest with pCoHygro (Invitrogen) and selecting in the presence of 100 μg/ml Hygromycin B (Enzo Life Sciences) for ~1 month. Transfections were performed with Cellfectin (Life Technologies), according to manufacturers’ procedures.
IR Treatments
Cultures were exposed to IR using a 160 kV X-ray source (X-RAD iR-160, Precision X-Ray). A dose of 5 Gy was used for the experiment, as in previous studies (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016, Caridi et al. 2018, Merigliano et al. 2023) .
Plasmids
pCopia-3xFlag-3xHA(FHA)-Nup153 FL or ∆C plasmids were generated by inserting the corresponding PCR-amplified sequence of Nup153 into a pCopia-FHA backbone (Chiolo et al. 2011, Ryu et al. 2015, Ryu et al. 2016, Caridi et al. 2018, Merigliano et al. 2023) , after AscI/PacI digestion. The cDNA template used was LD46479 from DGRC.
dsRNA synthesis and sequences
dsRNAs were prepared with the MEGAscript T7 Kit (Thermo Fisher Scientific Cat# AM1334). dsRNAs targeting the C-terminus of Nup153 were prepared using the oligos: Nup153 for: 5’-CTAATACGACTCACTATAGGGAG-CCCACACCTTTGTCGAACTT and Nup153 rev: 5’-CTAATACGACTCACTATAGGGAG-ACAGGCCACACACTTGTTGA.
RNAi depletions in cultured cells
dsRNAs were transfected with DOTAP Liposomal Transfection Reagent (Roche) following manufacturer’s instructions. Incubation times and dsRNA amounts were optimized to maximize depletion efficiency while avoiding toxicity and cell cycle effects. Cells were treated with dsRNAs for 5 days For Nup153 depletion. The control (Ctrl) used for all RNAi experiments is RNAi depletion of the brown (bw) gene transcript, which regulates the body color of adults flies and is not involved in DNA repair pathways (Chiolo et al. 2011, Ryu et al. 2015, Caridi et al. 2018, Merigliano et al. 2023) .
Western blotting
1-3×10 6 cells were collected, washed once in PBS and lysed for 15–20 min on ice with lysis buffer (50 mM Tris, pH 7.8, 1% NP-40, 150 mM NaCl) containing protease inhibitors (Complete, Roche), 2-mercaptoethanol, and 1 mM PMSF. Benzonase was added to each sample (0.5 U/ul) for 30 min. The soluble lysate was recovered by centrifugation (10 min, 4°C) and resuspended in loading buffer (Laemmli) to a final concentration of 1x. Samples were denatured for 5 min at 95°C before running them on a TGX 4–12% gel (Bio-Rad). Samples were then transferred onto nitrocellulose membrane for hybridization with specific antibodies.
Immunofluorescence and quantification of repair foci in fixed samples
Immunofluorescence was performed as previously described (Chiolo et al. 2011, Ryu et al. 2015, Caridi et al. 2018, Merigliano et al. 2023) . Imaging and image processing was performed with the DeltaVision Deconvolution microscope and the Softworx Software as previously described (Chiolo et al. 2011, Ryu et al. 2015, Caridi et al. 2018, Merigliano et al. 2023) . Classification of repair foci inside or outside the DAPI-bright region was done as in (Chiolo et al. 2011, Ryu et al. 2015, Caridi et al. 2018, Merigliano et al. 2023) .
Statistical analysis
All statistical analyses were performed using Prism6 (Graphpad Software).
Reagents
|
Antibodies |
|||
|
Name |
concentration |
source |
catalogue number |
|
Histone H2AvD phosphoS137 |
1:500 IF |
Rockland |
600-401-914 |
|
anti-Nup153 |
1:500 WB |
M. Capelson |
N/A |
|
anti-Nup107 |
1:2000 |
V. Doye |
N/A |
|
anti-HA |
1:1000 IF |
DSHB |
rMs-IgG1 |
|
anti-Flag |
1:1000 WB |
Sigma-Aldrich |
F1804 |
IF: immunofluorescence, WB: western blotting, DSHB: Developmental Studies Hybridoma Bank.
|
Plasmids name |
Source |
Stock # |
|
pCopia-FHA-Nup153 |
This study |
p155 |
|
pCopia-FHA-Nup153∆C |
This study |
p394 |
Acknowledgments
Acknowledgments
We thank V. Doye and M. Capelson for antibodies, Hannah Hopp and Melissa Bonner for generating plasmids, and B. Spatola for help with the blots.
Funding Statement
<p>This work was funded by an AAUW International Fellowship to C.M.; a USC Research Enhancement Fellowships to T. Ryu; NIH R01GM117376 and NSF Career 1751197 to I.C. The original Nup153 clone was obtained from DGRC (P40OD010949). Antibodies against HA were obtained from DSHB, created by the NICHD of the NIH and maintained at the University of Iowa.</p>
References
- Amaral N, Ryu T, Li X, Chiolo I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017 Jan 16;33(2):86–100. doi: 10.1016/j.tig.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A. 2003 Jan 27;100(3):981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, D'Agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature. 2018 Jun 20;559(7712):54–60. doi: 10.1038/s41586-018-0242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, Delabaere L, Tjong H, Hopp H, Das D, Alber F, Chiolo I. Quantitative Methods to Investigate the 4D Dynamics of Heterochromatic Repair Sites in Drosophila Cells. Methods Enzymol. 2018 Feb 26;601:359–389. doi: 10.1016/bs.mie.2017.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi CP, Plessner M, Grosse R, Chiolo I. Nuclear actin filaments in DNA repair dynamics. Nat Cell Biol. 2019 Sep 3;21(9):1068–1077. doi: 10.1038/s41556-019-0379-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caridi PC, Delabaere L, Zapotoczny G, Chiolo I. And yet, it moves: nuclear and chromatin dynamics of a heterochromatic double-strand break. Philos Trans R Soc Lond B Biol Sci. 2017 Oct 5;372(1731) doi: 10.1098/rstb.2016.0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011 Feb 25;144(5):732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Tang J, Georgescu W, Costes SV. Nuclear dynamics of radiation-induced foci in euchromatin and heterochromatin. Mutat Res. 2013 Aug 16;750(1-2):56–66. doi: 10.1016/j.mrfmmm.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dialynas G, Delabaere L, Chiolo I. Arp2/3 and Unc45 maintain heterochromatin stability in Drosophila polytene chromosomes. Exp Biol Med (Maywood) 2019 Jul 31;244(15):1362–1371. doi: 10.1177/1535370219862282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004 Jan 12;15(4):1991–2002. doi: 10.1091/mbc.e03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JW, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, Sohn KA, Minoda A, Tolstorukov MY, Appert A, Parker SC, Gu T, Kundaje A, Riddle NC, Bishop E, Egelhofer TA, Hu SS, Alekseyenko AA, Rechtsteiner A, Asker D, Belsky JA, Bowman SK, Chen QB, Chen RA, Day DS, Dong Y, Dose AC, Duan X, Epstein CB, Ercan S, Feingold EA, Ferrari F, Garrigues JM, Gehlenborg N, Good PJ, Haseley P, He D, Herrmann M, Hoffman MM, Jeffers TE, Kharchenko PV, Kolasinska-Zwierz P, Kotwaliwale CV, Kumar N, Langley SA, Larschan EN, Latorre I, Libbrecht MW, Lin X, Park R, Pazin MJ, Pham HN, Plachetka A, Qin B, Schwartz YB, Shoresh N, Stempor P, Vielle A, Wang C, Whittle CM, Xue H, Kingston RE, Kim JH, Bernstein BE, Dernburg AF, Pirrotta V, Kuroda MI, Noble WS, Tullius TD, Kellis M, MacAlpine DM, Strome S, Elgin SC, Liu XS, Lieb JD, Ahringer J, Karpen GH, Park PJ. Comparative analysis of metazoan chromatin organization. Nature. 2014 Aug 28;512(7515):449–452. doi: 10.1038/nature13415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, Wan KH, Park S, Mendez-Lago M, Rossi F, Villasante A, Dimitri P, Karpen GH, Celniker SE. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007 Jun 15;316(5831):1625–1628. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins RA, Carlson JW, Wan KH, Park S, Mendez I, Galle SE, Booth BW, Pfeiffer BD, George RA, Svirskas R, Krzywinski M, Schein J, Accardo MC, Damia E, Messina G, Méndez-Lago M, de Pablos B, Demakova OV, Andreyeva EN, Boldyreva LV, Marra M, Carvalho AB, Dimitri P, Villasante A, Zhimulev IF, Rubin GM, Karpen GH, Celniker SE. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015 Jan 14;25(3):445–458. doi: 10.1101/gr.185579.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Löbrich M, Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic Acids Res. 2011 Apr 21;39(15):6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Li Q, Tjong H, Li X, Gong K, Zhou XJ, Chiolo I, Alber F. The three-dimensional genome organization of Drosophila melanogaster through data integration. Genome Biol. 2017 Jul 31;18(1):145–145. doi: 10.1186/s13059-017-1264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merigliano, C., T. Ryu, C. D. See, C. P. Caridi, N. Butova, T. Reynolds, . . . I. Chiolo (2023). "'Off-pore' nucleoporins relocalize heterochromatic breaks through phase separation." bioRxiv: 2023.2012.2007.570729.
- Miller, J. M., S. Prange, H. Ji, A. R. Rau, V. Y. Khodaverdian, X. Li, . . . I. Chiolo (2023). "Alternative end-joining results in smaller deletions in heterochromatin relative to euchromatin." eLife: 2023.2003.2003.531058.
- Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PGS, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AFA, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JMD, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science. 2022 Mar 31;376(6588):44–53. doi: 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008 Mar 26;18(2):204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal CC, Caridi CP, Chiolo I. Actin' between phase separated domains for heterochromatin repair. DNA Repair (Amst) 2019 Jul 8;81:102646–102646. doi: 10.1016/j.dnarep.2019.102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, Gorchakov AA, Jaffe JD, Kennedy C, Linder-Basso D, Peach SE, Shanower G, Zheng H, Kuroda MI, Pirrotta V, Park PJ, Elgin SC, Karpen GH. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2010 Dec 22;21(2):147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Bonner MR, Chiolo I. Cervantes and Quijote protect heterochromatin from aberrant recombination and lead the way to the nuclear periphery. Nucleus. 2016 Sep 27;7(5):485–497. doi: 10.1080/19491034.2016.1239683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015 Oct 26;17(11):1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See C, Arya D, Lin E, Chiolo I. Live Cell Imaging of Nuclear Actin Filaments and Heterochromatic Repair foci in Drosophila and Mouse Cells. Methods Mol Biol. 2021;2153:459–482. doi: 10.1007/978-1-0716-0644-5_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsouroula K, Furst A, Rogier M, Heyer V, Maglott-Roth A, Ferrand A, Reina-San-Martin B, Soutoglou E. Temporal and Spatial Uncoupling of DNA Double Strand Break Repair Pathways within Mammalian Heterochromatin. Mol Cell. 2016 Jul 7;63(2):293–305. doi: 10.1016/j.molcel.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Vollmer B, Lorenz M, Moreno-Andrés D, Bodenhöfer M, De Magistris P, Astrinidis SA, Schooley A, Flötenmeyer M, Leptihn S, Antonin W. Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev Cell. 2015 Jun 4;33(6):717–728. doi: 10.1016/j.devcel.2015.04.027. [DOI] [PubMed] [Google Scholar]