Figure 2. Structural and sequence features of miRNA substrates and targets.
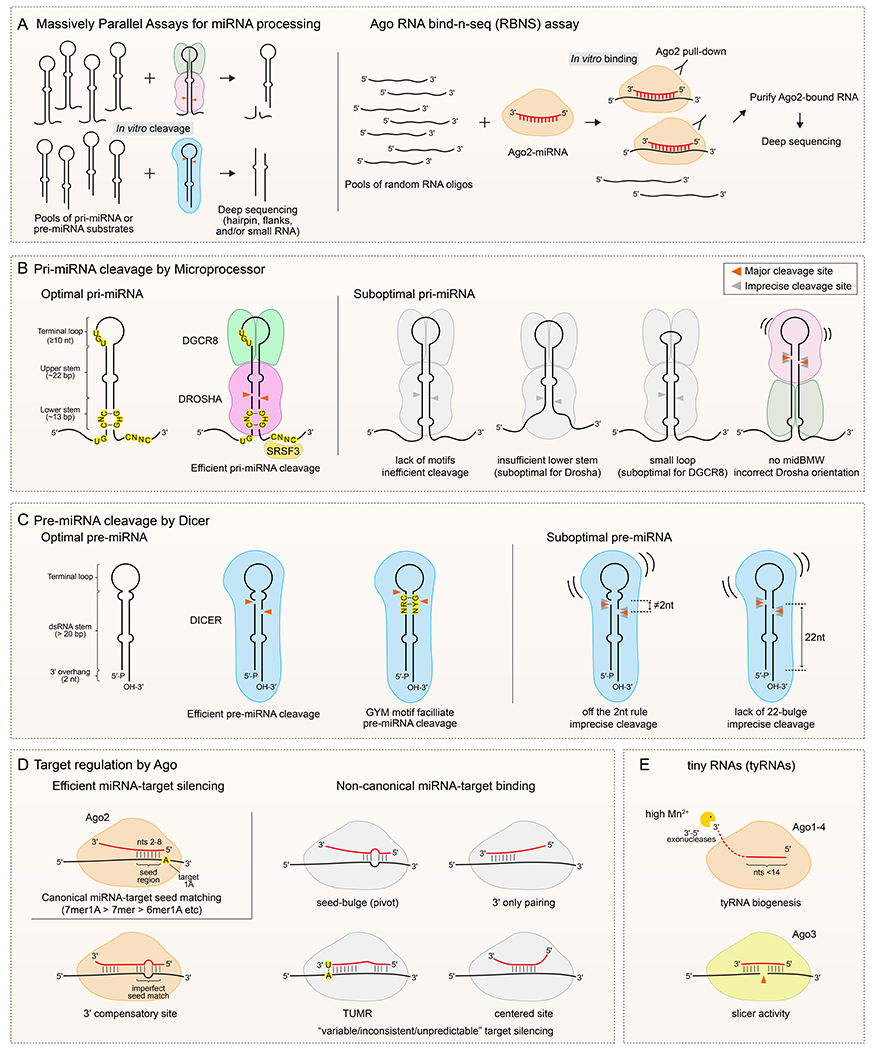
A. Design of massively parallel assays and RNA bind-n-seq (RBNS) assays for identifying miRNA pathway substrates and targets. (Left) A library of pri-miRNA or pre-miRNA substrates is incubated with purified Microprocessor (upper) or Dicer complex (lower) for in vitro cleavage, or for in vivo processing in cells. The reaction products are analysed by deep sequencing to infer structural features and sequence motifs within effective substrates. (Right) Target RNA oligo pools with random sequences are incubated with purified Ago2 loaded with a specific miRNA. Sequencing of bound species permits the relative affinities of different target sequences to be inferred. B. Pri-miRNA features. An optimal pri-miRNA contains characteristic structural features (single-stranded flanking regions, a ~35 bp dsRNA stem and ≥10 nt apical loop), primary motifs (including UG in the basal junction, UGU/GUG in the apical junction and CNNC in the 3’ flanking region), and chimeric structure-motif features such as the mismatched GHG (mGHG) in the lower stem. Drosha recognizes the basal junction, UG motif, and mGHG, and measures ~11 bp from the basal junction to cleave the dsRNA stem (red arrow heads). The DGCR8 dimer recognizes the apical loop and the UGU/GUG motif. Accessory factors can regulate microprocessing of pri-miRNA, including binding of CNNC by SRSF3. Pri-miRNAs with suboptimal features such as missing motifs, unpaired lower stem, and/or a small apical loop, exhibit inefficient and/or imprecise cleavage. Note that a middle bulge, mismatch and wobble base pair (midBMW) in the upper stem can repress unproductive cleavage by Microprocessor from the apical loop side. C. Pre-miRNA features. An optimal pre-miRNA hairpin contains 5’-monophosphate, 2 nt 3’-end overhang, a double-stranded stem of >20 bp and a single-stranded terminal loop. Dicer can measure its cleavage site from both 5’ and 3’ hairpin ends, although not all pre-miRNA substrates are engaged at both termini. A GYM (paired G, paired pyrimidine and mismatched C or A) motif near the Dicer cleavage site promotes pre-miRNA processing. A bulge/mismatch near the dicing sites enhances Dicer cleavage efficiency and/or accuracy, when the first nucleotide of the miRNA-3p species is 2 nt from the bulge/mismatch or when there is a 22-nt bulge in the pre-miRNA 3’ arm. Lack of this feature can cause Dicer to cut poorly and/or imprecisely, yielding multiple miRNA isoforms or lower small RNA levels. D. Ago–miRNA interaction features. (Left, top) The canonical miRNA-target interaction is mediated by seed pairing between nucleotides 2-8 (seed region) of miRNA with target RNA, and is often sufficient for measurable target repression. Shorter seed regions can also be functional, especially when followed by an adenosine opposite the first miRNA position (t1A). 7mer1A: seed 2-8 pairing with t1A; 7mer: seed 2-8 pairing; 6mer1A: seed 2-7 with t1A. (Left, bottom) A special class of target interaction with discontinuous seed pairing is compensated by extensive 3’ pairing. (Right) A variety of miRNA-target interactions lacking canonical seed pairing are found from in vivo Ago2-CLIP assays, in vitro Ago2-RBNS data, and bioinformatics; four types are summarized here. In general, the efficacy of these non-canonical sites is weak or controversial, and they were mostly assayed in vitro or in reporter systems. Unlike the exemplary 3’ compensatory let-7 site in lin-41, whose non-seed pairing is critical for C. elegans development, the impact of other non-canonical sites is mostly unknown. E. Tiny RNAs (tyRNAs). Under in vitro conditions of high manganese, several 3’-5’ exoribonucleases (ISG20, TREX1, and ERI1) can trim Ago-loaded miRNA down to <14 nt, and ISG20-mediated trimmed miRNA activates Ago3 cleavage of target mRNA.
