Figure 4. Strategies for the regulation of miRNA biogenesis.
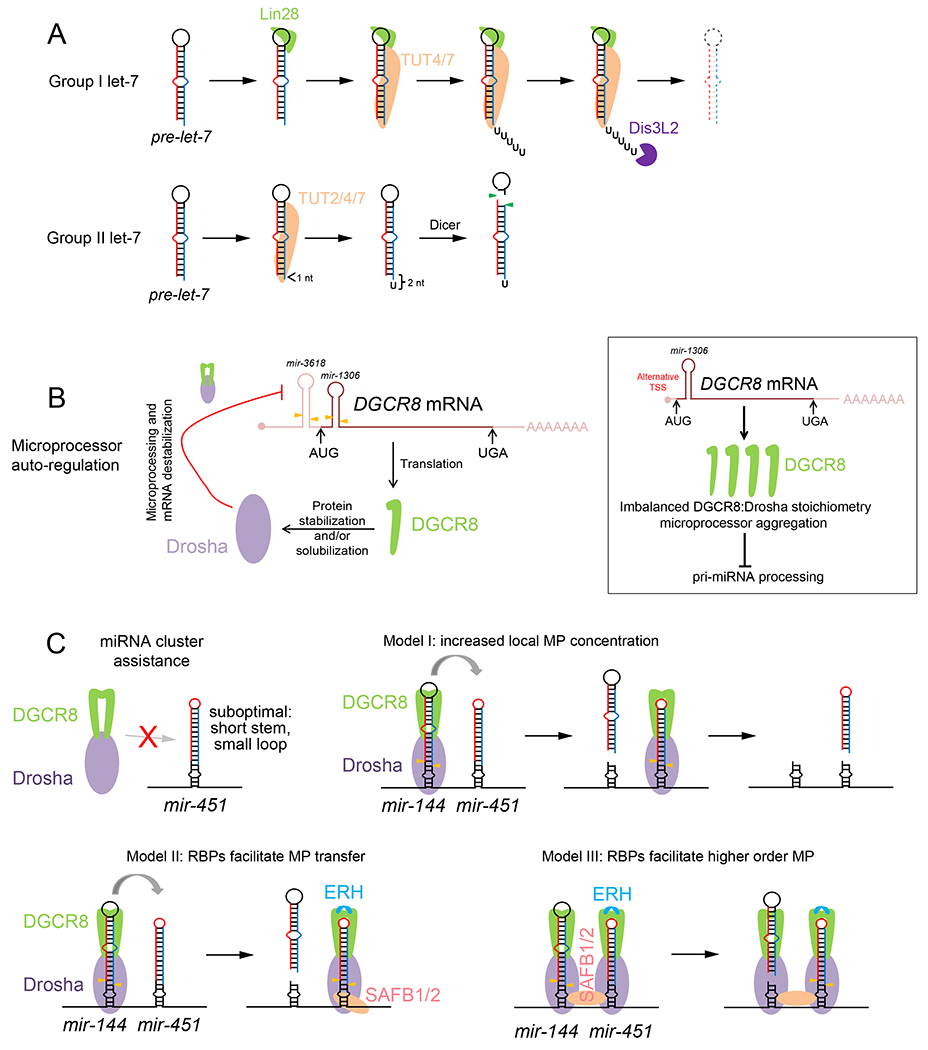
A. Two strategies of regulated maturation of let-7 miRNAs by 3’ tailing. With group I loci, Lin28 binds the terminal loop of pre-let-7 and recruits terminal uridyltransferases (TUT4/7) to oligouridylate its 3’ end, which triggers its degradation via Dis3L2. With group II loci, the initial Drosha-cleaved pre-let-7 only has a 1 nt-3’ overhang, rendering it a suboptimal Dicer substrate. However, this defect can be rescued by 3’-monouridylation via TUT2/4/7 to yield a 2 nt overhang, especially in absence of Lin28. B. Autoregulation of Microprocessor. (Left) The DGCR8 mRNA bears two hairpins within its 5’ UTR and CDS (mir-3618 and mir-1306), which can be cleaved by Drosha (yellow arrowheads). This destabilizes DGCR8 transcripts and reduces protein production. Reciprocally, DGCR8 protein positively regulates Microprocessor by stabilizing or solubilizing Drosha protein. (Right) However, during differentiation of mouse embryonic stem cells, usage of an internal promoter yields a shorter DGCR8 isoform lacking mir-3618, which escapes Microprocessor autoregulation. The resulting accumulation of DGCR8 imbalances DGCR8:Drosha protein stoichiometry, resulting in Microprocessor aggregation and reduced pri-miRNA processing. C. Enhanced biogenesis of suboptimal miRNAs within a genomic cluster. Certain miRNAs that lack optimal Microprocessor features (such as Dicer-independent mir-451) are poorly processed by themselves but are rescued by a neighbouring optimal miRNA (such as the normal context of mir-144/451 operon). Several models remain in play to explain this process, including increased local availability of Microprocessor via the miRNA helper, and various roles involving auxiliary RNA binding proteins (RBPs) such as ERH and SAFB1/2. These might stabilize Microprocessor on a suboptimal hairpin or bridge Microprocessor complexes via dimerization.
