Figure 5. Regulation of miRNA turnover.
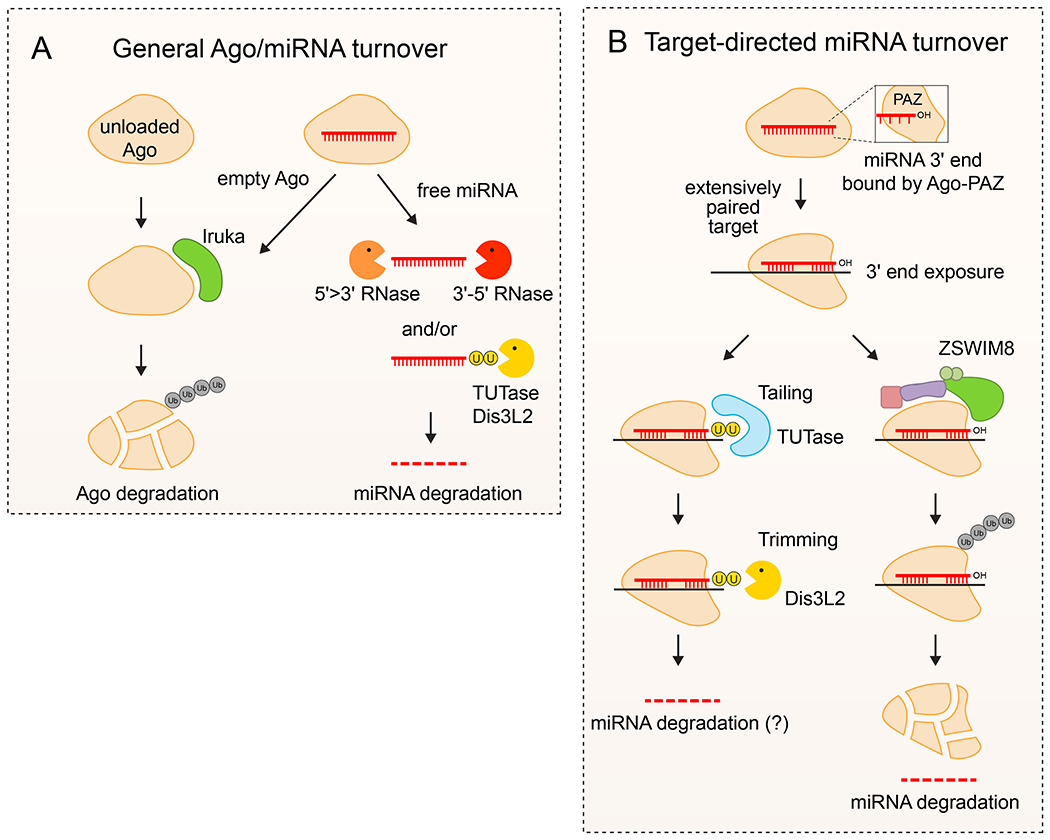
A. General turnover of Ago-miRNA complexes. Ago proteins that are not associated with small RNAs are usually unstable and selectively degraded across species. In principle, these exist prior to miRNA loading, or after ejection of the mature miRNA. Empty Ago proteins are selectively tagged by the E3 ubiquitin ligase Iruka, which signals its degradation by the proteasome. In concert, miRNAs are generally stable within Ago complexes. However, when not shielded by Ago protein, miRNAs are generally unstable since they lack 5’ cap and 3’ polyA tails. Such free miRNAs may be degraded directly by various exoribonucleolytic pathways or may be subject to 3’ tailing by TUTases and degradation by Dis3L2 3’ exoribonuclease. B. Target-directed miRNA degradation (TDMD) is a sequence-specific process by which individual designated miRNAs are degraded upon interaction with highly complementary trigger RNAs. Normally, the miRNA 3’-end is bound by the Ago-PAZ domain. However, when engaged in extensive base-pairing with a TDMD trigger, the miRNA 3’-end is exposed. This provides access to modification by TUTase and potential degradation by Dis3L2; however, tailing is not generally coupled to turnover. Instead, the dominant mechanism for TDMD involves a conformational change of Ago that allows it to be selectively recognized by the ZSWIM8, which recruits a Cullin-RING E3 ubiquitin ligase complex to degrade Ago. The exposed miRNA is then presumably subject to turnover by general exoribonucleases.
