Abstract
Cinnamomum tamala, commonly known as “Indian bay leaf” or “Tejpat”, is an economically important plant widely used in medicine, food and cosmetic industries. Growing demand for its leaf and bark in the herbal trade and non-availability of quality materials lead to large-scale species admixture and adulteration in the global market. The present study aims at developing a validated HPLC-DAD (High-performance liquid chromatography coupled with diode array detection) method and multiple markers-based chemical fingerprints for quality evaluation of C. tamala leaf extracts. Five bioactive compounds, viz., coumarin, cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate, were identified and quantified in 28 samples collected from the western Himalayan region of India. The chromatographic separation was achieved on Shimadzu Shimpak C18 column (dimension 250 × 4.6 mm, pore size 5 μm) with a gradient elution of mobile phase using acetonitrile and 0.1 percent phosphate buffer and the chromatograms were obtained at a wavelength of 265 nm. The method validation was done by analyzing the linearity, LOD, LOQ, precision, stability, repeatability and recovery rates of standard compounds for quantitative analysis. The values of coefficient of correlation (R2) were found to be close to 1 for linearity and similarity analysis; and standard deviation was less than 3 percent in case of precision, stability, repeatability and recovery rates. The content of target compounds such as coumarin, cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate varied in the range of 0–1.09, 0–0.05, 0.07–0.51, 0.39–1.27 and 0–0.27 percent, respectively. In the chemical fingerprint of C. tamala leaves, a total of 13 peaks were assigned as common peaks. The results of the study indicated that the HPLC method now developed combining chemical fingerprint with quantification of analytes could serve as a useful tool for quality evaluation of herbal raw materials of C. tamala and a valuable reference for further study.
Keywords: HPLC, Chemical fingerprint, Quality control, Herbal products, Cinnamomum tamala and medicinal plant
1. Introduction
The genus Cinnamomum (Lauraceae) is comprised of about 248 species of trees and shrubs, which are distributed in tropical and sub-tropical Asia to Western Pacific region. The bark and leaves of several Cinnamomum species contain aromatic essential oils; and are widely used as spices, flavouring agents and also in traditional and modern medicines [1]. C. verum (True cinnamon or Ceylon cinnamon), C. cassia (Chinese cinnamon), C. burmannii (Indonesian cinnamon), C. loureiroi (Vietnamese cinnamon) and C. tamala (Indian Cassia or Indian Bay leaf) are among the economically most important spe of this genus [2]. Of these, Cinnamomum tamala Nees and Eberm, known as Tajpata/Tamalapatra or Indian bay leaf, is native to the wet tropical forests of India, Bangladesh, Nepal, Bhutan, China, Laos, Myanmar, Vietnam and both Eastern and western Himalayas and also cultivated in many other tropical and sub-tropical countries of the world. The dried leaves and bark of C. tamala are being used since the first century A.D. in the Indian subcontinent as spices or flavouring agents in tea, savoury meals and traditional foods due to its spicy, peppery, clove-cinnamon-like flavour [3]. Ancient Greek and Roman literature mentioned about the trade of Indian bay leaf from Malabar (Kerala) coast, India under the trade name Malabathra or Malabathron [4]. In Indian system of medicine, the leaves are used against rheumatism, diarrhoea, colic pain, vomiting and nausea. The leaf essential oil is reported to exhibit antioxidant, antibacterial, antifungal, hypoglycaemic and other biological activities; and has potential applications against cancer, diabetes, cardiac diseases, anxiety, ulcer, depression and GI diseases [5,6]. Further, the pleasant aroma of the essential oil makes it suitable for use in perfumery and food processing industries [7]. The medicinal properties of C. tamala could be attributed to the presence of a wide range of bioactive compounds. The crude plant extract is reported to contain several alkaloids, reducing sugar, tannin, amino acids, glycosides, terpenoids, phenols, flavonoids (quercetin, kaempferol, and quercetrin), ascorbate and carotenoids, saponins and steroids [[8], [9], [10], [11]] Similarly, eugenol, methyl eugenol, cinnamaldehyde, trans-cinnamyl acetate etc. are the major constituents of the essential oil of C. tamala having a broad range of pharmacological activities [6,12,13].
During the past 3-4 decades, in view of the potential adverse effects of modern drugs, there has been a resurgence of alternative therapies based on plants and plant products worldwide and consequently, the demand of herbal products and supplements has increased many-fold [14]. Paradoxically, however, the unsustainable harvesting from the wild and destruction of the natural habitats have brought about large-scale depletion of medicinal plant resources [15].
With ever-increasing demand of raw materials of highly-traded medicinal plants, their scanty availability in the wild and limited cultivation in farmers' field, there is growing concern about the widespread adulteration and species admixtures in the herbal medicinal products [16]. Sometimes, the product adulteration could be due to misidentification or substitution with an allied congeneric species [17]. Lately, the adverse effects of species adulteration in the herbal trade on the health and safety of consumers have been realized and documented for few species [16]. The leaves of C. tamala are one of the high-value traded commodities in India's spice and pharmaceutical sectors. Frequently, the leaves of allied species of Cinnamomum such as C. malabatrum and C. verum are mixed with C. tamala leaves as adulterant and this unfair practice is still continuing in India and elsewhere [18]. The chemical fingerprinting has proved to be an efficient technique for the characterization of secondary metabolites such as phenolics, alkaloids, saponins, terpenes, lipids, carbohydrates etc. associated with the particular plant species and this can play a key role in quality control of plant materials used for herbal drugs [19]. The Food and Drug Administration, World Health Organization and the European Medicines Agency have recognized the chromatographic fingerprint as the most useful and effective method for authentication, quality control and consistency of herbal products [20]. There are a number of chromatographic and spectrophotometric techniques which are frequently used to ascertain the drug quality of herbal medicines. However, high performance liquid chromatography (HPLC) fingerprint has emerged as the most popular and widely used technique for authentication and qualitative evaluation of botanical products due to its simplicity and versatility for analysis in terms of separation, identification and quantification of chemical compounds [21,22].
The present investigation is aimed at optimizing the HPLC chromatographic conditions, validating the methods as per ICH (International Council for Harmonisation) guidelines and developing a HPLC chemical fingerprint using multiples markers for checking the adulteration and authentic identification of this highly traded medicinal plant Cinnamomum tamala collected from the Western Himalayan regions of India.
2. Materials and methods
2.1. Plant samples and chemical reagents
The leaves of C. tamala were collected from different locations of Himachal Pradesh (HP), Uttarakhand and Jammu and Kashmir (J & K) in the Western Himalayan region of India in the month of July 2022. The details of samples collected with the sample code, name of the locality, GPS coordinates and altitude are provided in Table 1. The leaves were collected from mature and healthy plants of approximately 30–50 m in height. The HPLC grade chemicals, which included methanol (≥99.9 %), acetonitrile (≥99.9 %), ortho-phosphoric acid (≥97 %) and water were procured from Merck Life Science Pvt. Ltd., Mumbai, India. The standards, which included coumarin (>99 %), cinnamyl alcohol (>98 %), cinnamaldehyde (>95 %) and cinnamyl acetate (>98 %) were procured from Sigma-Aldrich, Spruce Street, St. louis, USA and cinnamic acid (>99 %) procured from Merck KGaA, Darmstadt, Germany. The chemical structures of the compounds are shown in Fig. 1.
Table 1.
List of samples of C. tamala collected from different locations of Western Himalayan regions.
| Code | Name of the place | Latitude | Longitude | Altitude |
|---|---|---|---|---|
| CTL-1 | Huranga Narayan, HP | 31° 56′ 11.0004″ N | 76° 56′ 51″ E | 1844 |
| CTL-2 | Saroli, HP | 32° 0′ 20.9988″ N | 76° 44′ 30.0012″ E | 1310 |
| CTL-3 | Nagan, HP | 32° 0′ 19.0008″ N | 76° 44′ 17.0016″ E | 1310 |
| CTL-4 | Nagan, HP | 32° 0′ 6.0012″ N | 76° 44′ 8.0016″ E | 1310 |
| CTL-5 | Nagan, HP | 32° 0′ 6.0012″ N | 76° 44′ 8.0016″ E | 1310 |
| CTL-6 | Chauntra, HP | 32° 0′ 52.9992″ N | 76° 44′ 9.9996″ E | 1310 |
| CTL-7 | Mandi, HP | 31° 42′ 9.684″ N | 76° 56′ 31.092″ E | 759 |
| CTL-8 | Sheel, Pungapur, Mandi, HP | 31° 38′ 22.600″ N | 76° 48′ 29.900″ E | 1481 |
| CTL-9 | Hatgargh, Mandi, HP | 31° 32′ 29.688″ N | 76° 56′ 51.288″ E | 933 |
| CTL-10 | Nagan Bharola, Mandi, HP | 31° 59′ 48.912″ N | 76° 43′ 12.108″ E | 1192 |
| CTL-11 | Nauhli (Nohli), Mandi, HP | 31° 52′ 30.108″ N | 76° 50′ 55.716″ E | 1066 |
| CTL-12 | FTI, Sundarnagar, HP | 31° 31′ 32.304″ N | 76° 54′ 32.508″ E | 955 |
| CTL-13 | Sarohli, Lower Chuntra, HP | 32° 0′ 17.1″ N | 76° 43′ 46.2″ E | 1194 |
| CTL-14 | Khadi, J & K | 32° 38′ 59.82″ N | 75° 54′ 24.588″ E | 1086 |
| CTL-15 | Gatinala, J & K | 32° 41′ 15.828″ N | 75° 49′ 39.828″ E | 1246 |
| CTL-16 | Bastewal, J & K | 32° 48′ 22.392″ N | 75° 22′ 29.388″ E | 1005 |
| CTL-17 | Keya, J & K | 32° 46′ 9.66″ N | 75° 26′ 55.392″ E | 1148 |
| CTL-18 | Ranibagh, Uttarakhand | 29°17 ′4.00″ N | 79° 32′ 45.91″ E | 594 |
| CTL-19 | Bhujia Ghat - Uttarakhand | 29° 18′ 27.792″ N | 79° 31′ 41.592″ E | 725 |
| CTL-20 | Dolmar, Uttarakhand | 29° 19′ 12.828″ N | 79° 31′ 2.28″ E | 785 |
| CTL-21 | Jeoli Kot, Uttarakhand | 29° 20′ 39.84″ N | 79° 29′ 9.708″ E | 1163 |
| CTL-22 | Do Gaon, Uttarakhand | 29° 19′ 21.54″ N | 79° 30′ 14.22″ E | 922 |
| CTL-23 | Nalni, Uttarakhand | 29° 20′ 36.6″ N | 79° 22′ 27.732″ E | 1183 |
| CTL-24 | Jalal Gaon, Uttarakhand | 29° 22′ 19.272″ N | 79° 22′ 37.38″ E | 1273 |
| CTL-25 | Pasholi, Uttarakhand | 29° 16′ 48.252″ N | 79° 34′ 41.232″ E | 1057 |
| CTL-26 | Gumal Gaon, Uttarakhand | 29° 15′ 43.272″ N | 79° 35′ 32.532″ E | 1032 |
| CTL-27 | Paniya, Uttarakhand | 29° 15′ 12.24″ N | 79° 36′ 23.652″ E | 1098 |
| CTL-28 | Kaladungi, Uttarakhand | 29° 17′ 29.508″ N | 79° 20′ 21.048″ E | 388 |
Fig. 1.
Chemical structure of five standard compounds.
2.2. Sample and standard preparation
The leaf samples were pulverized into powder after being shade-dried. The powdered samples were run through a sieve of pore size 250 μm (BSS 60 mesh) and placed in a zipper-lock polythene bag for further analysis. The powdered samples were extracted using 50 percent methanol in HPLC-grade water. About 0.2 g of sample was dissolved in 20 ml of solvent. It was then sonicated for 30 min at 25-30 °C. The solution was then filtered through a 0.22 μm syringe filter and stored in HPLC vials. The filtrates were used for the HPLC analysis. The samples and extract preparation methods are shown in Fig. 2. The standard compounds like coumarin, cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate were dissolved in methanol with the starting concentration of 180, 380, 124, 98 and 100 mg/L respectively. These stock solutions were gradually diluted and injected into HPLC in order to get calibration curves for quantification purposes.
Fig. 2.
Processing plant samples and preparation of plant extract: A: C. tamala, B: Dried leaf sample, C: powder plant sample, D: Ultrasonic extraction, E: Filtration of solution with 0.22μ membrane filtration, F: Plant extract for HPLC analysis.
2.3. HPLC chromatographic conditions
The samples were then subjected to HPLC analysis for quantitative estimation of bioactive compounds. The components of the Shimadzu HPLC system (Kyoto, Japan) utilized for HPLC analysis were a CBM-20A controller, a CT0-20AC column oven, an SPD-20A diode array detector, a binary LC-20 AD pump and a Rheodyne 8125 injector. Using solvents, A (acetonitrile) and B (0.1 % phosphate buffer in HPLC grade water) as mobile phases with a gradient flow from pump B which was 90 percent for 0–12 min, 80 percent for 13–35 min, 50 percent for 36–40 min and 0 percent for 40–60 min the chromatographic separation was done in a Shimadzu Shimpak C18 column having dimension of 250 × 4.6 mm and 5 μm pore size. The flow rate was maintained at 1 ml/min. The standard solutions and sample injection volumes were both set at 20 μl. The run took 60 min in total. The PDA detector was set up at 265 nm for obtaining chromatograms and the UV spectra were captured between 190 and 800 nm. After obtaining the chromatograms of the sample and standard compounds, the retention time and absorption spectra of the target peaks were recorded to identify the compounds present in the plant samples. Then, the compounds in the samples were quantified by using standard calibration curves.
2.4. Method validation and quantitative analysis of bioactive compounds
The validity of the method was tested to check the stable interaction of the compounds towards the stationary and mobile phases. The method validation of quantitative analysis was conducted for linearity, accuracy, stability, repeatability, limit of detection (LOD) and limit of quantification (LOQ) as well as recovery test, in compliance with the guidelines of the International Conference on Harmonization [23]. The analyte concentration at which the signal peak area is at least three times greater than the signal to noise ratio is known as the LOD. The limit of quantitation (LOQ) is the lowest concentration of an analyte at which the peak area is greater than the signal-to-noise ratio (S/N) by a factor of at least ten. To determine the precision, the replicate solution of each standard was injected four times in a single day. The same solution of each standard was injected for four days in a row to determine the standard's stability (0, 24, 48 and 72 h). Four replicates of each reference standard solution were prepared independently in order to assess each compound's repeatability. The recovery test was ascertained through the application of the standard addition method, which involved reanalysing the sample after a certain quantity of standard was added. The peak area of the previously analysed sample and standard were added, which were regarded as the expected peak area in the recovery study. The peak area calculated after injecting the standard and sample mixture constitutes the observed peak area for the calculation of the recovery percentage. The bioactive compounds were identified and quantified using the developed calibration curves.
2.5. Development of chemical fingerprint
The chromatographic fingerprints were developed by taking the chromatogram of selected samples having good resolution and peak separation. The peaks that appeared invariably in all the samples were regarded as common characteristic peaks. The common characteristic peaks include the peaks of known compounds and others were regarded as unknown peaks. The establishment of a reference chromatogram assisted in the determination of the similarity indices among the samples. The development of the reference chromatogram involved a comparison between the retention time and peak area of every sample chromatogram and the average of these parameters amongst all samples. The correlation between the sample and reference chromatograms was determined by calculating the relative standard deviation (RSD) of the relative retention time (RRT) and relative peak area (RPA).
2.6. Validation for HPLC fingerprint
The developed chemical fingerprint was also validated to check the stable interaction of the compounds in the plant samples with the chromatographic phases. The validation was done following the Chinese State Food and Drug Administration regulations [24] and taking one of the studied samples, i.e. leaf sample (CTL- 4). The precision, consistency and repeatability of common peaks in the sample chromatogram were considered for validating the HPLC fingerprint. The precision test involved preparation of the sample solution and analysing it four times, successively. The stability was examined by injecting the same sample solution (CTL – 4) at different time points (0 h, 24 h, 48 h and 72 h), after preparation. By injecting biological replicates of the sample solution, the repeatability of the sample was assessed. For the repeatability analysis, four replicates were injected. The result was expressed as the RSD of the RRT and RPA of the identified common peaks in relation to the reference peak. Thirteen (13) common peaks selected in the present study and peak 12 (cinnamaldehyde) was selected as the reference (R) peak, having the highest peak area in all the samples as compared to peak areas of other common peaks of the chemical fingerprint.
3. Results and discussion
3.1. Selection of HPLC method and identification of marker compounds
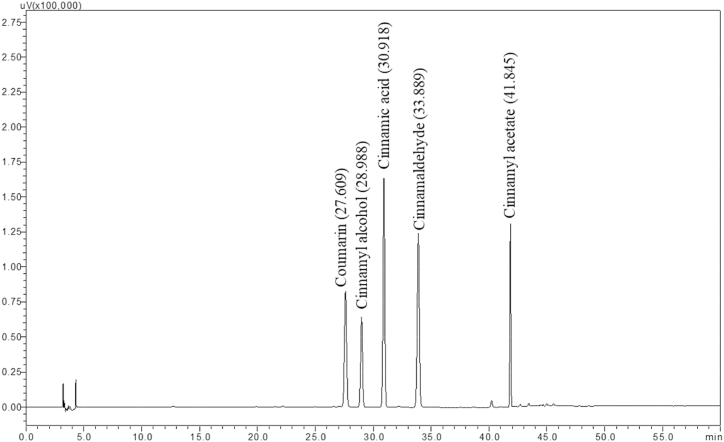
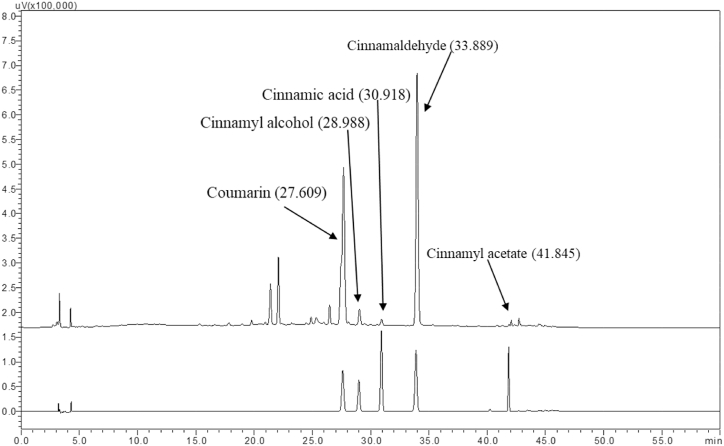
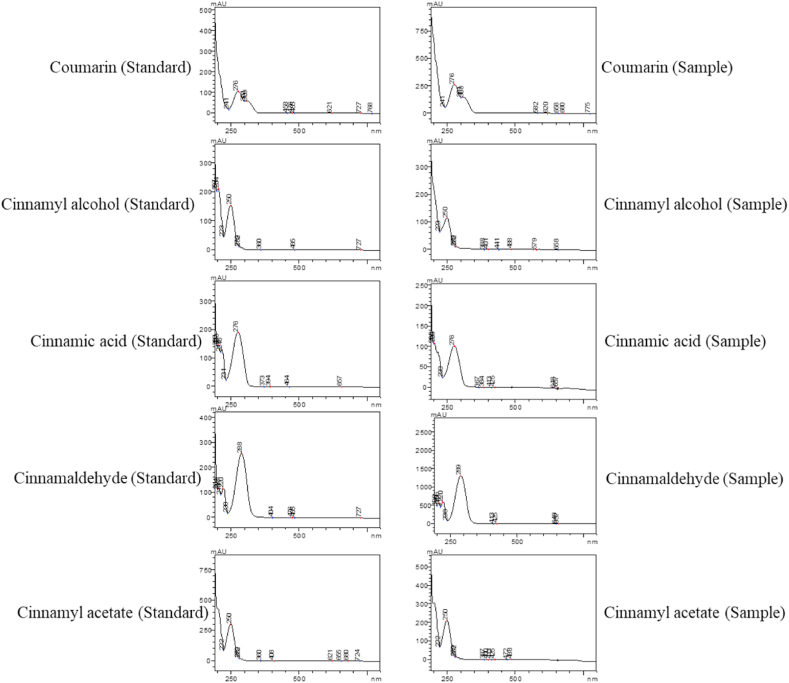
In the present study, the HPLC method was optimized for the quality assessment of C. tamala samples collected from various geographical locations of Western Himalayan region. In view of the fact that the marker profiling and standardization are very crucial for enumerating phytoconstituents of the medicinal plants [25], a total of five chemical marker compounds were taken for the analysis. The method was developed by critically analysing different chromatographic methods used for the separation of compounds in the related species of the same genus Cinnamomum [26,27], including C. cassia [28,29]. The method described by Ding et al. [29] was found to be suitable for the current work and was used in the study with slight modification in total run time. This method was selected based on elution of greater number and highly resolved peaks and complete elution of all compounds in comparison to other methods tried in this study. The method was found to be highly efficient in separation and identification of standard compounds and common peaks. In order to further validate the chromatographic parameters, different mobile phases, such as acetonitrile or methanol and water containing phosphate and acetic buffer were tested. It was found that acetonitrile and phosphate buffer solution offered a more stable baseline, with more peaks compared to other mobile phases. To improve the peak shape and restrain the peak tailing, the concentration and pH value of phosphate buffer aqueous solution were investigated. Three concentrations of phosphate buffer (0.1 %, 0.2 % and 0.3 %) were used. The findings suggest that the optimal elution was acetonitrile and 0.1 percent phosphate buffer aqueous solution (pH 3.0 adjusted with orthophosphoric acid) as it had the best peak shapes and baseline resolution. The gradient used in the current study can be considered a ‘scouting gradient’ that involves the online (dynamic) mixing of solvents to achieve a steady increase in the elution strength of the organic solvent (acetonitrile) from a low value to a high value over a run time period of 60 min. The HPLC chromatograms of the standard compounds are given in Fig. 3. The comparative chromatogram identifying the peaks of the corresponding standard compounds in the samples are given in Fig. 4. The spectral comparison between the target peaks in standard and sample chromatograms are given in Fig. 5. The spectral analysis gives a stronger evidence with regard to the presence of these compounds in the plant sample along with the retention time.
Fig. 3.
Chromatogram of five standard compounds of C. tamala.
Fig. 4.
Comparison between the peaks of standard present in the sample near the same retention time.
Fig. 5.
Spectral comparison of target compounds in standard solution chromatogram and sample chromatogram.
Though there are few reports on the chromatographic analysis of some species of the genus Cinnamomum, no HPLC method has been optimized for C. tamala using five standards, simultaneously. The content of eugenol, isoeugenol and cinnamaldehyde was measured in leaf and bark samples by an LC method developed in C. tamala and C. zeylanicum [26]. They have concluded that this approach is simple and precise and may be applied to the quantitative estimation of eugenol, isoeugenol and cinnamaldehyde in these two Cinnamomum species. A reverse-phase HPLC method for simultaneous quantification of cinnamaldehyde and methyl eugenol in the methanolic leaf extract of C. zeylanicum has been developed [30]. The study concluded that this method is simple and precise which is suitable for quality evaluation of C. zeylanicum herbal products [30]. Similarly, another reverse-phase HPLC method for the quantitative determination of eugenol from the leaf extract of C. tamala and its polyherbal formulation has been standardized [12], which was found to be an appropriate method for accurate quantitative analysis of eugenol from C. tamala leaves and different pharmaceutical formulations prepared out of it. The chemical fingerprint analysis in Cinnamomum species has been done by taking seven bioactive constituents such as protocatechuic acid, epicatechin, cinnamic acid, cinnamaldehyde, eugenol and benzoic acid and the compounds have been identified and quantified using a HPLC method [27]. In order to differentiate the bark and twig samples of C. cassia collected from different geographical regions of China, Vietnam and Indonesia, HPLC fingerprint method has been successfully employed [29]. They found this method to be quite efficient in separating different components of the plant sample and in discriminating the chemotypes within a species.
Therefore, keeping in view of efficiency of HPLC method for quantitative and qualitative analysis of the bioactive compounds in Cinnamomum species, the present study presents a novel approach of optimizing HPLC method in C. tamala leaf samples for concurrent measurement of five key biologically active compounds for quality assessment. Importantly, the samples were taken from the native habitat of C. tamala located in a specific biogeographic zone i.e., Western Himalayan region, which ensures the purity of the samples in terms of its climatic parameters and genetic characteristics.
3.2. Method validation of quantitative analysis
The linearity of the calibration curve was found to be good for all the standard compounds and the linear regression equations were obtained. The calibration curves of all the five compounds are provided in Fig. 6. The coefficient of correlation of all the calibration curves were found to be between 0.984 and 0.995 (R2). The LOD and LOQ values of the compounds have shown that all these compounds can be detected and quantified even at a relatively low concentrations in the plant samples. Thus, the large variability in the concentration of chemical compounds among the plant samples would not affect the detection and quantification process in the optimized HPLC method. The details of values of linear range, regression equation, R2, LOD and LOQ of five standard components are given in Table 2. The precision, stability and recovery tests were done to check whether the standards of the same concentration are appearing at same Retention Time (RT) and showing the same Peak Area (PA) or not. The results were interpreted by calculating the peak area and retention time. The variations were recorded in terms of relative standard deviation among the RT and PA among the replicates (Table 3). In the precision test, the RSD percentage of RT and PA between the replicates of all the standard compounds were found to be in the range of 0.049–0.218 and 0.821–1.115, respectively. The stability test was found to have RSD percentage of 0.035–0.205 among RT and 0.931–1.704 among the PA of standard compounds. In the test of repeatability, these values varied in the range of 0.131–0.673 among RT and 0.839–1.200 among PA. The study of recovery was done to check the complete elution of the compound at the particular RT from the stationary phase. The recovery percentage of all the five compounds in our study were more than 98 percent, showing almost complete elution in the present HPLC method (Table 4). The value of coefficient of correlation (R2) close to 1 and the values of standard deviation less than 3 percent reported in the present analysis are considered as admissible ranges in statistical perspective.
Fig. 6.
Calibration Curves of five standard compounds for quantitative analysis.
Table 2.
Linear range, regression equation, R2, LOD and LOQ of five standard components.
| Component | Linear range (mg/L) | Regression equation | R2 | LOD (mg/L) | LOQ (mg/L) |
|---|---|---|---|---|---|
| Coumarin | 11.22–180 | y = 78.045x + 0.378 | 0.995 | 0.004 | 0.015 |
| Cinnamyl alcohol | 23.76–380 | y = 24.976x + 0.308 | 0.990 | 0.011 | 0.035 |
| Cinnamic acid | 7.75–124 | y = 181.01x + 0.649 | 0.994 | 0.004 | 0.012 |
| Cinnamaldehyde | 6.125–98 | y = 211.36x + 0.611 | 0.994 | 0.002 | 0.008 |
| Cinnamyl acetate | 6.25–100 | y = 84.929x + 0.485 | 0.984 | 0.006 | 0.019 |
Table 3.
Analytical results of precision, stability and repeatability of five standard compounds.
| Component | Precision (n = 4) |
Stability (n = 4) |
Repeatability (n = 4) |
|||
|---|---|---|---|---|---|---|
| RSD % of RT | RSD % of PA | RSD % of RT | RSD % of PA | RSD % of RT | RSD % of PA | |
| Coumarin | 0.218 | 1.115 | 0.205 | 1.256 | 0.673 | 1.200 |
| Cinnamyl Alcohol | 0.154 | 0.821 | 0.150 | 1.346 | 0.476 | 0.841 |
| Cinnamic Acid | 0.174 | 0.913 | 0.161 | 1.704 | 0.262 | 0.839 |
| Cinnamaldehyde | 0.185 | 1.097 | 0.168 | 1.049 | 0.308 | 1.051 |
| Cinnamyl Acetate | 0.049 | 0.648 | 0.035 | 0.931 | 0.131 | 0.677 |
Table 4.
Recovery rates of the five standard compounds of C. tamala in terms of peak area.
| Component | Original (Peak Area) | Found (Peak Area) | Recovery (%) | Average Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| Coumarin | 7369560 | 7082028 | 96.098 | 95.945 | 0.239 |
| 7083373 | 96.117 | ||||
| 7041947 | 95.555 | ||||
| 7075466 | 96.009 | ||||
| Cinnamyl alcohol | 5096542 | 5011181 | 98.325 | 98.618 | 0.175 |
| 5033821 | 98.769 | ||||
| 5029275 | 98.680 | ||||
| 5030232 | 98.699 | ||||
| Cinnamic acid | 12043908 | 12026152 | 99.853 | 99.982 | 0.181 |
| 12079173 | 100.293 | ||||
| 12029711 | 99.882 | ||||
| 12031698 | 99.899 | ||||
| Cinnamaldehyde | 14796026 | 14746935 | 99.668 | 99.509 | 0.198 |
| 14756248 | 99.731 | ||||
| 14685733 | 99.255 | ||||
| 14704629 | 99.382 | ||||
| Cinnamyl acetate | 4417139 | 4752529 | 107.593 | 107.178 | 0.275 |
| 4740454 | 107.320 | ||||
| 4722367 | 106.910 | ||||
| 4721406 | 106.888 |
The primary objective of quantitative and qualitative analyses through chromatographic techniques was to generate reliable, accurate and interpretable information about the active components in pharmaceutical samples. The analytical approach is put through a loosely defined validation study to make sure that it achieves this goal. Since the validation process is essential, particularly in trade, conflict situations and regulatory control, the chemical analysis ought to be clear and comprehensible, as far as possible [31]. Previously, in realization of this fact, researchers have done validation of their chromatographic methods in order to represent the results of analytical data in a robust and holistic manner [29,[32], [33], [34], [35], [36], [37], [38], [39], [40]]. Therefore, validation of optimized HPLC method in the present study signifies the standard requirement and authenticity of the chromatographic method.
The validated method was used for analysis of all the samples for identification and quantification of target compounds. A wide range of variation in the content of target compounds among the samples was observed. The amount of target compounds i.e., coumarin, cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate varied in the range of 0.000–1.094, 0.000–0.0542, 0.0705–0.5090, 0.3880–1.2703 and 0.000–0.2721 percent, respectively (Table 5). The quantitative data showed that the major bioactive compound of C. tamala was cinnamaldehyde, which was detected in comparatively high concentrations as compared with other quantified compounds. Though coumarin was not identified or quantified in any other samples, the samples from Uttarakhand were found to contain coumarins in higher quantities. The zero values in the content table for cinnamyl alcohol and cinnamyl acetate depicts extremely low concentration of compounds, which lies beyond the third decimal point of the quantified value. The zero value in the coumarin showed its absence in the samples mostly from Himachal Pradesh and Jammu & Kashmir. However, the value zero for coumarin in the Uttarakhand sample i.e., CTL-18 revealed that the concentration determined is below the quantification limit of the method in the current study although the peak has been detected. The absence or low concentration of coumarin in the samples are good for human use, because higher concentrations coumarin are considered hepatotoxic to human body [41]. Therefore, coumarin may act as a negative marker in the fingerprinting analysis as well as qualitative and quantitative analysis and its complete absence or low concentration in the samples would indicate the toxicity level of the samples.
Table 5.
Estimation of five bioactive compounds in C. tamala leaves through validated HPLC method.
| Sample code | Name of the place | Name of the State | Coumarin (%) | Cinnamyl alcohol (%) | Cinnamic acid (%) | Cinnamaldehyde (%) | Cinnamyl acetate (%) |
|---|---|---|---|---|---|---|---|
| CTL-1 | Huranga Narayan | Himachal Pradesh | 0.0000 | 0.0191 | 0.5090b | 1.2703b | 0.2213 |
| CTL-2 | Saroli | Himachal Pradesh | 0.0000 | 0.0000 | 0.2549 | 0.5741 | 0.0000 |
| CTL-3 | Nagan | Himachal Pradesh | 0.0000 | 0.0000 | 0.4016 | 0.8897 | 0.0312 |
| CTL-4 | Nagan | Himachal Pradesh | 0.0000 | 0.0000a | 0.1907 | 0.6634 | 0.0000a |
| CTL-5 | Nagan | Himachal Pradesh | 0.0000 | 0.0065 | 0.1790 | 0.7250 | 0.0823 |
| CTL-6 | Chauntra | Himachal Pradesh | 0.0000 | 0.0105 | 0.1631 | 0.9327 | 0.0255 |
| CTL-7 | Mandi, HP | Himachal Pradesh | 0.0000 | 0.0108 | 0.1489 | 0.4295 | 0.0313 |
| CTL-8 | Sheel, Pungapur, Mandi | Himachal Pradesh | 0.0000 | 0.0296 | 0.0935 | 0.4174 | 0.2579 |
| CTL-9 | Hatgargh, Mandi | Himachal Pradesh | 0.0000 | 0.0291 | 0.1419 | 0.6917 | 0.2392 |
| CTL-10 | Nagan Bharola, Mandi | Himachal Pradesh | 0.0000 | 0.0080 | 0.2840 | 0.7917 | 0.0153 |
| CTL-11 | Nohli, Mandi | Himachal Pradesh | 0.0000 | 0.0304 | 0.1376 | 1.0318 | 0.0747 |
| CTL-12 | FTI, Sundarnagar | Himachal Pradesh | 0.0000 | 0.0114 | 0.1949 | 0.5506 | 0.1250 |
| CTL-13 | Lower Chuntra | Himachal Pradesh | 0.0000 | 0.0122 | 0.2897 | 0.8128 | 0.0629 |
| CTL-14 | Khadi | J and K | 0.0000 | 0.0278 | 0.0966 | 0.6197 | 0.1541 |
| CTL-15 | Gatinala | J and K | 0.0000 | 0.0542b | 0.0839 | 0.5482 | 0.1723 |
| CTL-16 | Bastewal | J and K | 0.0000 | 0.0344 | 0.2205 | 0.6714 | 0.2721b |
| CTL-17 | Keya | J and K | 0.0000 | 0.0143 | 0.0919 | 0.6220 | 0.0607 |
| CTL-18 | Ranibagh | Uttarakhand | 0.0000a | 0.0111 | 0.2160 | 0.5228 | 0.0581 |
| CTL-19 | Bhujia Ghat | Uttarakhand | 0.0692 | 0.0076 | 0.1206 | 1.2277 | 0.2274 |
| CTL-20 | Dolmar | Uttarakhand | 0.0777 | 0.0169 | 0.2341 | 0.4818 | 0.0612 |
| CTL-21 | Jeoli Kot | Uttarakhand | 0.1774 | 0.0169 | 0.1471 | 0.7902 | 0.0382 |
| CTL-22 | Do Gaon | Uttarakhand | 1.0949b | 0.0239 | 0.0705a | 0.3880a | 0.0207 |
| CTL-23 | Nalni | Uttarakhand | 0.1639 | 0.0159 | 0.2418 | 0.6445 | 0.0702 |
| CTL-24 | Jalal Gaon | Uttarakhand | 0.0291 | 0.0091 | 0.4187 | 1.1559 | 0.0146 |
| CTL-25 | Pasholi | Uttarakhand | 0.6870 | 0.0061 | 0.1116 | 0.4902 | 0.0000 |
| CTL-26 | Gumal Gaon | Uttarakhand | 0.0283 | 0.0089 | 0.3293 | 0.8178 | 0.0209 |
| CTL-27 | Paniya | Uttarakhand | 0.0566 | 0.0339 | 0.0886 | 0.5517 | 0.0241 |
| CTL-28 | Kaladungi | Uttarakhand | 0.0288 | 0.0096 | 0.3712 | 0.7380 | 0.0543 |
Minimum value in the series.
Maximum value in the series, J & K- Jammu and Kashmir.
The variation of secondary metabolites is attributed to a number of internal and external factors. The quantity and quality of plant secondary metabolites may vary in response to fluctuations in seasonal climatic parameters and environmental factors [42]. The developmental stage of the plant and genetic factors also play key roles in secondary metabolite production and accumulation [43]. In the present study, since the samples were collected in a single season and the plants selected were of nearly equal age, it is most likely that the variation in bioactive compounds might not be due to changes in seasonal parameters and stages of plant development. Since the samples were gathered from different altitudes ranging from 388 m to 1844 m, the variability in secondary metabolites might be due to altitudinal variation [44,45]. Besides, the variation of secondary metabolites in C. tamala samples might be due to the combined effect of altitudinal variation and genetic factors, which needs precise validation in future studies.
Recent research indicates that differences in chemical diversity within species can influence community diversity among different trophic interactions [46,47]. Furthermore, it is crucial to explore and understand the natural range of variation to identify superior plant genotypes with desired traits. This inherent variability is vital for successful breeding efforts to develop high-yielding progenies. The diversity based on secondary metabolite content offers promising opportunities for selecting industrially profitable elite germplasm of C. tamala with high cinnamaldehyde content.
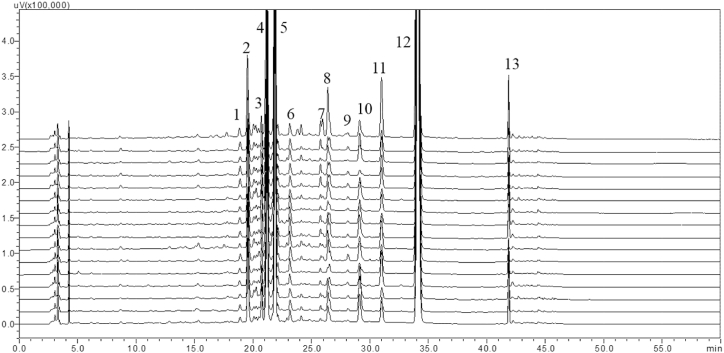
3.3. Development of chemical fingerprint and its validation
A total of 13 common peaks were identified in the fingerprint of selected samples. The chemical fingerprint developed for C. tamala is provided in Fig. 7. The peaks from 1 to 9 are of unidentified compounds and the peaks 10, 11, 12 and 13 represent cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate, respectively. Among them, the shape of common peak 12 was symmetrical and had the highest intensity (cinnamaldehyde). So, the peak 12 (cinnamaldehyde) was used as a reference peak to calculate the RRT and RPAs. The formulae for calculating RRT and RPAs were RRT = RT peak/RT peak12 and RPA = PA peak/PA peak12, respectively. The purpose of calculating RRT and RPAs was to make the various absolute values become relatively stable. The peak of coumarin was not taken into consideration because it does not occur in all the samples. This may be due to absent of these compounds in some samples or concentration of this compound was below the level of detection and quantification by HPLC method. As the limit of detection and quantification covers a wide range of concentrations, it could be assumed that the compound is not present in some samples. The similarity of all the samples were measured by comparing the RT and PA with the reference chromatogram which was developed by taking the average of RT and PA of each common peaks of all the samples taken for fingerprint study. The samples selected for fingerprint analysis were randomly named as S1 to S16. The similarity index in terms of coefficient of correlation (R2) with the reference chromatogram, was found to be more than 0.903. The detail values are given in Table 6. The R2 values were close to 1 in all the samples, which establishes their similarity with the refence chromatogram. The relationships among the common peaks with respect to a prominent and high intensity peak was found out for a more credible conclusion. The reference peak was selected based on the highest peak area and high resolution in all the samples. The peak no 12 i.e., cinnamaldehyde was the reference peak for calculating the RRT and RPA of each sample. The RSD among the relative retention time of the samples were found to be less than 0.266 percent signifying strong similarity among the RRT of the samples. However, the RSD of peak areas were found to be very high; the highest RSD being 188 percent. The high RSD among the peak areas of the samples signifies that the peak area in all the samples varied greatly due to presence of particular compound in varying concentrations (Table 7). The values of similarity index, RSD values of the relative retention time and peak area makes this chemical fingerprint a highly credible one. The stability of the developed fingerprint in the present HPLC method was then checked by validation study.
Fig. 7.
HPLC fingerprint of C. tamala leaf samples.
Table 6.
Similarity among the selected samples for fingerprint development with respect to reference chromatogram in terms of Retention Time and Peak Area of common peaks.
| Similarity index in terms of Peak Area | Similarity index in terms of Retention Time | ||
|---|---|---|---|
| Sample | Similarity | Sample | Similarity |
| S1 | 0.999 | S1 | 0.999 |
| S2 | 0.991 | S2 | 0.998 |
| S3 | 0.989 | S3 | 0.999 |
| S4 | 0.999 | S4 | 0.997 |
| S5 | 0.995 | S5 | 0.995 |
| S6 | 0.998 | S6 | 0.999 |
| S7 | 0.957 | S7 | 0.993 |
| S8 | 0.903 | S8 | 0.990 |
| S9 | 0.999 | S9 | 0.999 |
| S10 | 0.998 | S10 | 0.997 |
| S11 | 0.993 | S11 | 0.995 |
| S12 | 0.999 | S12 | 0.999 |
| S13 | 0.992 | S13 | 0.996 |
| S14 | 0.992 | S14 | 0.997 |
| S15 | 0.992 | S15 | 0.998 |
| S16 | 0.999 | S16 | 0.995 |
Table 7.
Retention time (RT), Relative retention time (RRT), Peak area (PA) and Relative peak area (RPA) of 13 characteristics fingerprint peaks in C. tamala (N = 16).
| Peak | Component | Average RT | RRT |
Average PA | RPA |
||
|---|---|---|---|---|---|---|---|
| Average | RSD (%) | Average | RSD (%) | ||||
| 1 | Unknown | 18.877 | 0.554 | 0.162 | 81751.125 | 0.006 | 34.606 |
| 2 | Unknown | 19.557 | 0.574 | 0.162 | 771568.938 | 0.059 | 50.684 |
| 3 | Unknown | 20.744 | 0.609 | 0.148 | 255486.688 | 0.020 | 53.388 |
| 4 | Unknown | 21.203 | 0.622 | 0.129 | 2384250.750 | 0.184 | 48.804 |
| 5 | Unknown | 21.877 | 0.642 | 0.131 | 4514668.375 | 0.348 | 41.931 |
| 6 | Unknown | 23.151 | 0.679 | 0.114 | 176640.500 | 0.014 | 46.772 |
| 7 | Unknown | 25.934 | 0.761 | 0.187 | 33969.500 | 0.003 | 188.137 |
| 8 | Unknown | 26.432 | 0.775 | 0.264 | 243355.813 | 0.019 | 61.320 |
| 9 | Unknown | 28.111 | 0.825 | 0.062 | 39352.063 | 0.003 | 29.007 |
| 10 | Cinnamyl alcohol | 29.125 | 0.854 | 0.037 | 272712.250 | 0.021 | 93.578 |
| 11 | Cinnamic acid | 31.008 | 0.910 | 0.024 | 393479.500 | 0.030 | 35.273 |
| 12 | Cinnamaldehyde (R) | 34.089 | 1.000 | 0.000 | 12976848.313 | 1.000 | 0.000 |
| 13 | Cinnamyl acetate | 41.876 | 1.228 | 0.056 | 343443.125 | 0.026 | 96.456 |
The HPLC chemical fingerprint was validated to check the stable interaction and affinity of the common peaks towards both the phases and parameters of the optimized HPLC method. The RSD between the RRT and RPA of all the common peaks of a samples in precision, stability and repeatability analysis were calculated and compared (Table 8). The RSD values of all the tests between RRT and RPA were below 2.952 percent, which suggest that the stability of these peaks is very strong towards the optimized HPLC method.
Table 8.
Analytical results of precision, stability and repeatability of 13 characteristics peaks in C. tamala sample (CTL-4).
| Peak no. | RSD of RRT (%) |
RSD of RPA (%) |
||||
|---|---|---|---|---|---|---|
| Precision | Stability | Repeatability | Precision | Stability | Repeatability | |
| 1 | 1.543 | 0.137 | 0.046 | 2.311 | 2.601 | 2.792 |
| 2 | 1.041 | 0.102 | 0.024 | 0.308 | 1.167 | 1.950 |
| 3 | 0.422 | 0.125 | 0.015 | 1.701 | 2.421 | 2.888 |
| 4 | 0.334 | 0.115 | 0.011 | 1.163 | 1.812 | 2.609 |
| 5 | 0.260 | 0.129 | 0.015 | 0.545 | 1.237 | 2.163 |
| 6 | 0.087 | 0.141 | 0.023 | 2.707 | 0.843 | 2.794 |
| 7 | 0.130 | 0.126 | 0.017 | 0.516 | 1.683 | 1.973 |
| 8 | 0.038 | 0.075 | 0.012 | 2.290 | 2.019 | 2.587 |
| 9 | 0.063 | 0.089 | 0.007 | 2.740 | 1.366 | 2.646 |
| 10 | 0.081 | 0.043 | 0.010 | 2.111 | 0.979 | 1.983 |
| 11 | 0.001 | 0.026 | 0.008 | 2.009 | 2.580 | 2.952 |
| 12 (R) | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
| 13 | 0.143 | 0.097 | 0.012 | 1.535 | 2.856 | 2.731 |
The chemical fingerprint is a widely adopted approach that has been used for the purpose of quality assessment, authentic identification and checking the adulteration and species admixtures in raw herbal drugs as well as marketed products [20,27,[48], [49], [50]] He et al. [51] developed HPLC chemical fingerprint for quality evaluation and quantitative analysis of tea by taking 10 batches of samples. A reproducible HPLC chemical fingerprinting approach has also been found out for the assessment of quality and consistency of Liuwei Dihuang Pills produced by different manufacturers in China [52]. The study revealed that along with quality assessment, this method could quantify the presence of some important constituents also.
Therefore, the HPLC chemical fingerprint approach is an efficient tool for the quality evaluation of medicinal plants and herbal products. The result of the present study on development of chemical fingerprint of C. tamala samples is a novel one and of contemporary relevance, which would be useful for quality evaluation of this highly traded and industrially important medicinal plant.
4. Conclusion and future scope
The leaves of Cinnamomum tamala are among the most traded and exported high-value plant products meeting the needs of food and pharmaceutical industries globally. Due to non-availability of authentic materials or wrong identification of the species or ignorance, the leaves of Cinnamomum malabatrum, C. verum, Laurus nobilis (Bay leaf) etc. with different phytoconstituents and bioactivities are used as adulterants or admixtures leading to different nutritional compositions and pharmacological effects. In the present work, a reliable and efficient HPLC-DAD method combining HPLC fingerprint and quantitative analysis of major bioactive compounds has been developed and optimized for quality control of Cinnamomum tamala leaves for the first time. A characteristic HPLC fingerprint has been successfully established with simultaneous content determination of five bioactive constituents such as coumarin, cinnamyl alcohol, cinnamic acid, cinnamaldehyde and cinnamyl acetate. The method validation parameters show that the developed HPLC method is precise, accurate and robust for analysis of C. tamala leaf samples for quality assessment in terms of bioactive constituents. The HPLC chemical fingerprint developed in the present study would be of great significance for preventing adulteration in the herbal products and species admixture. In view of the tremendous economic potential of C. tamala leaves for food and medicine sectors, the methods developed and information generated from this study would be of great significance for authentic identification and quality evaluation of this highly traded and medicinally important plant.
Funding
This work is financially supported by Department of Biotechnology, Ministry of Science and Technology, Govt. of India (Grant No. BT/PR45247/NER/95/1926/2022).
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Bibhuti Bhusan Champati: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Prabhat Kumar Das: Writing – original draft, Investigation, Formal analysis, Data curation, Conceptualization. Chiranjibi Sahoo: Writing – original draft, Formal analysis, Data curation, Conceptualization. Asit Ray: Methodology, Investigation. Sudipta Jena: Methodology, Investigation. Ambika Sahoo: Methodology, Investigation. Sanghamitra Nayak: Writing – review & editing, Supervision. Swaran Lata: Project administration. Pratap Chandra Panda: Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
All authors declare that they have no conflict of interest.
Acknowledgement
The authors express their gratitude to S.C. Si, Dean, School of Pharmaceutical Sciences, Centre of Biotechnology and M.R. Nayak, President, Siksha 'O' Anusandhan (deemed to be University) for their assistance and encouragement.
References
- 1.Rao P.V., Gan S.H. Cinnamon: a multifaceted medicinal plant. Evid.-Based Complement. Alternat. Med. 2014;2014:1–12. doi: 10.1155/2014/642942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baruah A., Nath S.C. In: Cinnamon and Cassia-The Genus Cinnamomum. first ed. Ravindran P.N., Nirmal Babu K., Shylaja M., editors. CRC Press; Boca Raton, Florida, USA: 2004. Indian Cassia; pp. 215–226. [Google Scholar]
- 3.Tiwari S., Talreja S. Importance of Cinnamomum tamala in the treatment of various diseases. Phcog. J. 2020;12:1792–1796. doi: 10.5530/pj.2020.12.241. [DOI] [Google Scholar]
- 4.Sharma V., Rao L.J.M. An overview on chemical composition, bioactivity and processing of leaves of Cinnamomum tamala. Crit. Rev. Food Sci. Nutr. 2014;54:433–448. doi: 10.1080/10408398.2011.587615. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A.K., Mishra A. Antifungal and antioxidative potential of oil and extracts derived from leaves of Indian spice plant Cinnamomum tamala. Cell. Mol. Biol. 2012;58:142–147. [PubMed] [Google Scholar]
- 6.Kumar S., Vasudeva N., Sharma S. GC-MS analysis and screening of antidiabetic, antioxidant and hypolipidemic potential of Cinnamomum tamala oil in streptozotocin induced diabetes mellitus in rats. Cardiovasc. Diabetol. 2012;11:95. doi: 10.1186/1475-2840-11-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kapoor I.P.S., Singh B., Singh G. Essential oil and oleoresins of Cinnamomum tamala (Tejpat) as natural food preservatives for pineapple fruit juice. J. Food Process. Preserv. 2008;32:719–728. doi: 10.1111/j.1745-4549.2008.00200.x. [DOI] [Google Scholar]
- 8.Ahmed J., Sultana N., Dewan S.M., Amin M.N., Uddin S.N. Determination of chemical groups and investigation of anthelmintic, cytotoxic, and antibacterial activities of leaves of Cinnamomum tamala (Family: Lauraceae) Int. J. Pharmamedix India. 2013;1:222–232. [Google Scholar]
- 9.Prasad K.N., Yang B., Dong X., Jiang G., Zhang H., Xie H., Jiang Y. Flavonoid contents and antioxidant activities from Cinnamomum species. Innovat. Food Sci. Emerg. Technol. 2009;10:627–632. doi: 10.1016/j.ifset.2009.05.009. [DOI] [Google Scholar]
- 10.Mishra A.K., Singh B.K., Pandey A.K. In vitro-antibacterial activity and phytochemical profiles of Cinnamomum tamala (Tejpat) leaf extracts and oil. Reviews in Infection. 2010;1:134–139. [Google Scholar]
- 11.Chakraborty U., Das H. Antidiabetic and antioxidant activities of Cinnamomum tamala leaf extracts in STZ-treated diabetic rats. Glob. J. Biotechnol. Biochem. 2010;5:12–18. [Google Scholar]
- 12.Dighe V.V., Gursale A.A., Sane R.T., Menon S., Patel P.H. Quantitative determination of eugenol from Cinnamomum tamala Nees and Eberm. leaf powder and polyherbal formulation using reverse phase liquid chromatography. Chromatographia. 2005;61:443–446. doi: 10.1365/s10337-005-0527-6. [DOI] [Google Scholar]
- 13.Rana V.S., Langoljam R.D., Verdeguer M., Blázquez M.A. Chemical variability in the essential oil of Cinnamomum tamala L. leaves from India. Nat. Prod. Res. 2012;26:1355–1357. doi: 10.1080/14786419.2011.599806. [DOI] [PubMed] [Google Scholar]
- 14.Sen S., Chakraborty R., De B. Challenges and opportunities in the advancement of herbal medicine: India's position and role in a global context. J. Herb. Med. 2011;1:67–75. doi: 10.1016/j.hermed.2011.11.001. [DOI] [Google Scholar]
- 15.Chen S.L., Yu H., Luo H.M., Wu Q., Li C.F., Steinmetz A. Conservation and sustainable use of medicinal plants: problems, progress, and prospects. Chin. Med. 2016;11:1–10. doi: 10.1186/s13020-016-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srirama R., Santhosh Kumar J.U., Seethapathy G.S., Newmaster S.G., Ragupathy S., Ganeshaiah K.N., Uma Shaanker R., Ravikanth G. Species adulteration in the herbal trade: causes, consequences and mitigation. Drug Saf. 2017;40:651–661. doi: 10.1007/s40264-017-0527-0. [DOI] [PubMed] [Google Scholar]
- 17.Newmaster S.G., Grguric M., Shanmughanandhan M., Ramalingam S., Ragupathy S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013;11:222–235. doi: 10.1186/1741-7015-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Kumar K.N.S., Rajalekshmi M., Sangeetha B., Ravishankar B., Muralidhar R. Chemical examination of leaves of Cinnamomum malabatrum (Burm. f.) Blume sold as Tamalapatra. Pharmacogenomics J. 2012;4:11–15. doi: 10.5530/pj.2012.31.3. [DOI] [Google Scholar]
- 19.Syed M., Khan M.N., Khadim A., Shadab H., Perveen A., El-Seedi H.R., Musharraf S.G. Chemical fingerprinting of three Anemone species and an adulteration study to detect cross mixing of medicinal plants by HPLC-HR-ESI-MS/MS method. J. King Saud Univ. Sci. 2021;33 [Google Scholar]
- 20.Champati B.B., Jena S., Ray A., Padhiari B.M., Haldar T., Mohanty S., Sahoo A., Kar B., Ghosh B., Nayak S. Quality control and discrimination of Andrographis paniculata (Burm. f.) Nees based on High Performance Liquid Chromatography fingerprinting combined with chemometric approaches. Indian J. Pharma. Sci. 2021;83:1129–1143. doi: 10.36468/pharmaceutical-sciences.868. [DOI] [Google Scholar]
- 21.Liang Y.Z., Xie P., Chan K. Quality control of herbal medicines. J. Chromatogr. B. 2004;812:53–70. doi: 10.1016/j.jchromb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 22.Mukherjee P.K. first ed. Business Horizons Limited; New Delhi, India: 2002. Quality Control of Herbal Drugs: an Approach to Evaluation of Botanicals. [Google Scholar]
- 23.Guidance for Industry: Q2B Validation of Analytical Procedures, Methodology. U.S. Dept. Of Health and Human Services, Food and Drug Administration. Center for Drug Evaluation and Research: Center for Biologics Evaluation and Research; Rockville, MD, USA: 1996. [Google Scholar]
- 24.State Food and Drug Administration Bureau of China . 2002. Guidance for Experimental Research on HPLC Fingerprint of Traditional Chinese Injections (Draft) Beijing. [Google Scholar]
- 25.Leu Y.L., Kuo S.M., Hwang T.L., Chiu S.T. The inhibition of superoxide anion generation by neutrophils from Viscum articulatum. Chem. Pharm. Bull. 2004;52:858–860. doi: 10.1248/cpb.52.858. [DOI] [PubMed] [Google Scholar]
- 26.Dighe V.V., Gursale A.A., Charegaonkar G.A. Quantitation of eugenol, cinnamaldehyde and isoeugenol from Cinnamomum tamala nees and Eberm. Leaf powder and Cinnamomum zeylanicum breyn stem bark powder by LC. Chromatographia. 2009;70:1759–1762. doi: 10.1365/s10337-009-1382-7. [DOI] [Google Scholar]
- 27.Yang J., Chen L.H., Zhang Q., Lai M.X., Wang Q. Quality assessment of Cortex cinnamomi by HPLC chemical fingerprint, principal component analysis and cluster analysis. J. Separ. Sci. 2007;30:1276–1283. doi: 10.1002/jssc.200600389. [DOI] [PubMed] [Google Scholar]
- 28.Herbal Medicines Compendium, Cinnamomum Cassia Twig, US Pharmacopeia (https://hmc.usp.org./family/cinnamomum-cassia).
- 29.Ding Y., Wu E.Q., Liang C., Chen J., Tran M.N., Hong C.H., Jang Y., Park K.L., Bae K., Kim Y.H., Kang J.S. Discrimination of cinnamon bark and cinnamon twig samples sourced from various countries using HPLC-based fingerprint analysis. Food Chem. 2011;127:755–760. doi: 10.1016/j.foodchem.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Gursale A., Dighe V., Parekh G. Simultaneous quantitative determination of cinnamaldehyde and methyl eugenol from stem bark of Cinnamomum zeylanicum Blume using RP-HPLC. J. Chromatogr. Sci. 2010;48:59–62. doi: 10.1093/chromsci/48.1.59. [DOI] [PubMed] [Google Scholar]
- 31.Jenke D.R. Chromatographic method validation: a review of current practices and procedures. I. General concepts and guidelines. J. Liq. Chromatogr. Relat. Technol. 1996;19:719–736. doi: 10.1080/10826079608005533. [DOI] [Google Scholar]
- 32.Chandra A., Rana J., Li Y. Separation, identification, quantification, and method validation of anthocyanins in botanical supplement raw materials by HPLC and HPLC−MS. J. Agric. Food Chem. 2001;49:3515–3521. doi: 10.1021/jf010389p. [DOI] [PubMed] [Google Scholar]
- 33.Natividade M.M.P., Correa L.C., de Souza S.V.C., Pereira G.E., de Oliveira Lima L.C. Simultaneous analysis of 25 phenolic compounds in grape juice for HPLC: method validation and characterization of Sao Francisco Valley samples. Microchem. J. 2013;110:665–674. doi: 10.1016/j.microc.2013.08.010. [DOI] [Google Scholar]
- 34.Silva J.A., Borges N., Santos A., Alves A. Method validation for cafestol and kahweol quantification in coffee brews by HPLC-DAD. Food Anal. Methods. 2012;5:1404–1410. doi: 10.1007/s12161-012-9387-5. [DOI] [Google Scholar]
- 35.Rajauria G. Optimization and validation of reverse phase HPLC method for qualitative and quantitative assessment of polyphenols in seaweed. J. Pharm. Biomed. Anal. 2018;148:230–237. doi: 10.1016/j.jpba.2017.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Ramireddy A.R., Behara D.K. Analytical method validation on simultaneous estimation of Ozenoxacin and Benzoic acid in pharmaceutical formulation. Acta Chromatogr. 2023;35:278–285. doi: 10.1556/1326.2022.01064. [DOI] [Google Scholar]
- 37.Lee H.D., Paje L.A., Choi J., Kim J., Yu A.R., Bae M.J., Lee S. HPLC/DAD method validation of 6-hydroxyluteolin 7-O-glucoside analysis from Salvia plebeia. Korean J. Pharmacogn. 2021;52:186–191. doi: 10.22889/KJP.2021.52.3.186. [DOI] [Google Scholar]
- 38.Mehta S., Sharma A.K., Singh R.K. Method validation for the simultaneous estimation of three-bioactive components in combined extracts of three hepatoprotective plants using RP-HPLC method. J. Appl. Pharmaceut. Sci. 2021;11:127–131. [Google Scholar]
- 39.Mahrouse M.A., Lamie N.T. Experimental design methodology for optimization and robustness determination in ion pair RP-HPLC method development: application for the simultaneous determination of metformin hydrochloride, alogliptin benzoate and repaglinide in tablets. Microchem. J. 2019;147:691–706. doi: 10.1016/j.microc.2019.03.038. [DOI] [Google Scholar]
- 40.Asteggiano A., Curatolo L., Schiavo V., Occhipinti A., Medana C. Development, validation, and application of a simple and rugged HPLC method for boswellic acids for a comparative study of their abundance in different species of Boswellia gum resins. Appl. Sci. 2023;13:1254. doi: 10.3390/app13031254. [DOI] [Google Scholar]
- 41.Loncar M., Jakovljevic M., Subaric D., Pavlic M., Buzjak Sluzek V., Cindric I., Molnar M. Coumarins in food and methods of their determination. Foods. 2020;9:645. doi: 10.3390/foods9050645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soni U., Brar S., Gauttam V.K. Effect of seasonal variation on secondary metabolites of medicinal plants. Int. J. Pharma Sci. Res. 2015;6:3654–3662. doi: 10.13040/IJPSR.0975-8232.6(9).3654-62. [DOI] [Google Scholar]
- 43.Li Y., Kong D., Fu Y., Sussman M.R., Wu H. The effect of developmental and environmental factors on secondary metabolites in medicinal plants. Plant Physiol. Biochem. 2020;148:80–89. doi: 10.1016/j.plaphy.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Zidorn C. Altitudinal variation of secondary metabolites in flowering heads of the Asteraceae: trends and causes. Phytochemistry Rev. 2010;9:197–203. doi: 10.1007/s11101-009-9143-7. [DOI] [Google Scholar]
- 45.Pandey G., Khatoon S., Pandey M.M., Rawat A.K.S. Altitudinal variation of berberine, total phenolics and flavonoid content in Thalictrum foliolosum and their correlation with antimicrobial and antioxidant activities. J. Ayurveda Integr. Med. 2018;9:169–176. doi: 10.1016/j.jaim.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glassmire A.E., Jeffrey C.S., Forister M.L., Parchman T.L., Nice C.C., Jahner J.P., Wilson J.S., Walla T.R., Richards L.A., Smilanich A.M., Leonard M.D. Intraspecific phytochemical variation shapes community and population structure for specialist caterpillars. New Phytol. 2016;212:208–219. doi: 10.1111/nph.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richards L.A., Lee A.D., Matthew L.F., Angela M.S., Craig D.D., Michael D.L., Christopher S.J. Phytochemical diversity drives plant–insect community diversity. Proc. Natl. Acad. Sci. USA. 2015;112:10973–10978. doi: 10.1073/pnas.150497711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y., Wu T., Zhu J., Wan L., Yu Q., Li X., Cheng Z., Guo C. Combinative method using HPLC fingerprint and quantitative analyses for quality consistency evaluation of an herbal medicinal preparation produced by different manufacturers. J. Pharm. Biomed. Anal. 2010;52:597–602. doi: 10.1016/j.jpba.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Padhiari B.M., Ray A., Halder T., Jena S., Champati B.B., Sahoo A., Kar B., Ghosh B., Panda P.C., Nayak S. Quality control of Bacopa monnieri by High-Performance Liquid Chromatography fingerprinting combined with chemometric methods. Phcog. Mag. 2021;17:872–881. doi: 10.4103/pm.pm_107_21. [DOI] [Google Scholar]
- 50.Springfield E.P., Eagles P.K.F., Scott G. Quality assessment of South African herbal medicines by means of HPLC fingerprinting. J. Ethnopharmacol. 2005;101:75–83. doi: 10.1016/j.jep.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 51.He X., Li J., Zhao W., Liu R., Zhang L., Kong X. Chemical fingerprint analysis for quality control and identification of Ziyang green tea by HPLC. Food Chem. 2015;171:405–411. doi: 10.1016/j.foodchem.2014.09.026. [DOI] [PubMed] [Google Scholar]
- 52.Xie B., Gong T., Tang M., Mi D., Zhang X., Liu J., Zhang Z. An approach based on HPLC-fingerprint and chemometrics to quality consistency evaluation of Liuwei Dihuang Pills produced by different manufacturers. J. Pharm. Biomed. Anal. 2008;48:1261–1266. doi: 10.1016/j.jpba.2008.09.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.