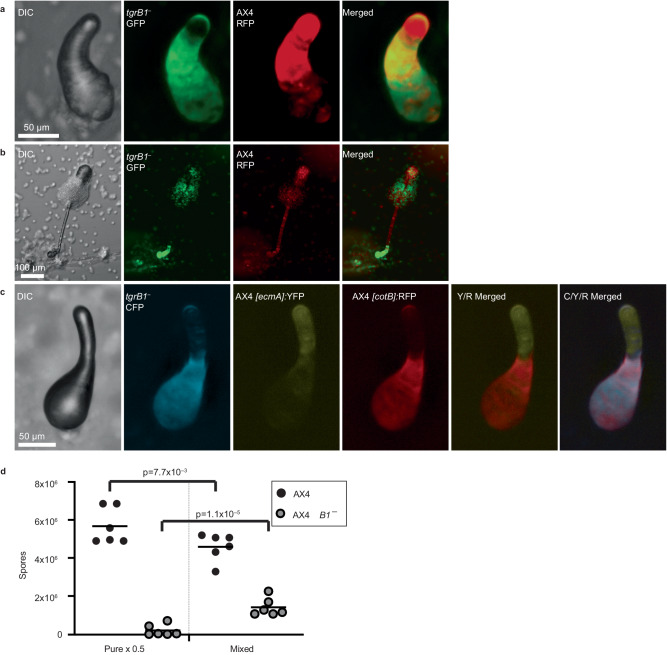
Fig. 2. Inactivation of tgrB1 confers cheating behavior.
We used two strains that express constitutive fluorescent markers, the wild-type AX4-RFP (red) and the inactivated tgrB1 mutant AX4 tgrB1–-GFP (green). We grew the cells separately, mixed equal proportions, and co-developed them. We imaged the structures at the finger stage (a) and fruiting body stage (b) with DIC and with green and red fluorescence, and generated a merged image of the red and green channels, as indicated. The green staining at the edge of the anterior region is due to reflection and is not associated with cells. c We also mixed constitutively labeled mutant tgrB1–-CFP cells (cyan) with wild-type AX4 cells carrying the prestalk reporter [ecmA]:YFP (yellow) and the prespore reporter [cotB]:RFP (red). We imaged the structures with DIC and with cyan, yellow, and red fluorescence, and generated merged images of the yellow and red (Y/R) as well as all three channels (C/Y/R), as indicated. d We grew the cells separately, developed 7 × 106 cells either in pure populations or mixed at equal proportions as indicated, and counted spores. The spore counts are shown as six independent replicates (symbols) and their averages (horizontal lines). In this case, we used 4 different alleles of AX4 tgrB1–-GFP mixed with AX4-RFP and one of AX4 tgrB1–-RFP mixed with AX4-GFP in 6 experiments. The pure population counts were multiplied by 0.5 to scale them with the mixed population. Brackets and p-values (T-test, one sided, n = 6) compare the spore counts of each strain in the two conditions. Camera settings are included in Supplementary Data 1. Source data are provided as a Source Data file.