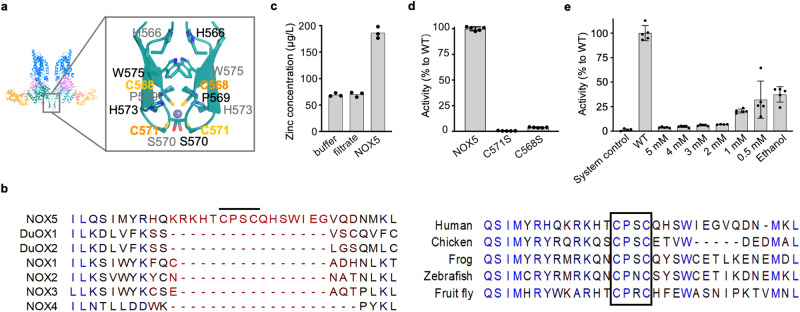
Fig. 4. The putative zinc binding site at the dimer interface.
a Structural details of the zinc binding site. b Sequence alignment of the CXXC-containing insertion between NOX5 paralogs and orthologs. The CXXC motif is indicated with black line and black box in left and right panel, respectively. c ICP-MS shows the presence of Zinc ions in NOX5. In x-axis: buffer indicates NOX5 sample buffer (20 mM Tris, 200 mM NaCl, 0.05% Digitonin); filtrate indicates the buffer passed through the filter device during NOX5 concentration; NOX5 is the experimental group. Data are shown with n = 3 technical repeats. Source data are provided as a Source Data file. d Relative activity of C571S and C568S to wild-type NOX5. Y-axis indicates the relative activity compared with NOX5 wild type. Data are shown (average +/-SD) with n = 5 biologically independent samples. Source data are provided as a Source Data file. e Dose-dependent inhibition of NOX5 by TPEN. Y-axis indicates the relative activity compared with NOX5 wild type. Data are shown with (average +/-SD) n = 5 biologically independent samples. Data are processed with GraphPad Prism (Version 10.1.1). We performed Grubbs’ test59 (also known as extreme studentized deviate, ESD), to determine the outliers. We excluded one data point in “2 mM TPEN” group, which is a significant outliner (P < 0.05) based on Grubbs’ test. Source data are provided as a Source Data file.