Figure 2. Deconstruction of fibroblast heterogeneity at E12.5 (expression and location).
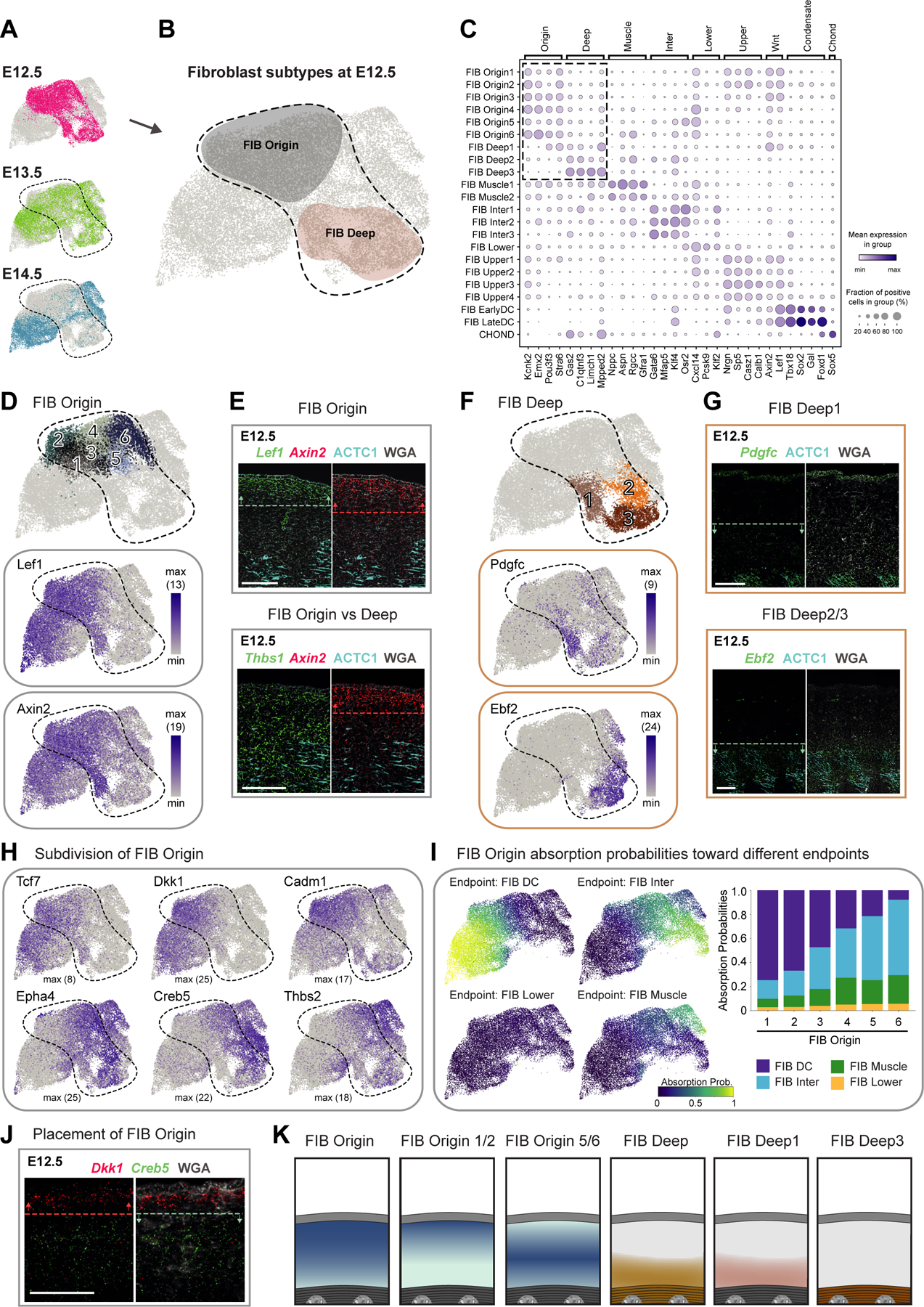
(A) UMAP visualization of fibroblasts from the different embryonic ages (n = 10 008 cells of E12.5, 8 016 of E13.5, and 7 920 of E14.5).
(B) Major fibroblast subtypes at E12.5 highlighted on UMAP.
(C) Marker gene expression dot plot for major fibroblast groups. Highlighted are the clusters mostly present at E12.5.
(D,F) Subclustering of early fibroblast groups (upper panels). Expression pattern of characteristic marker genes (lower panels). Number in brackets shows max number of RNA copies detected per cell (absolute abundance).
(E,G,J) mRNA (italics) and protein (capitalized) stainings of fibroblast subpopulations. Dashed lines and arrows highlight the region with highest expression. Microscope images originate from larger tile scans (n = 3 mice). Scale bars, 100μm.
(H) Marker gene expression of FIB Origin subpopulations on UMAP.
(I) Absorption probabilities towards the differentiation endpoints projected onto UMAP (left panels) and quantified for each FIB Origin subpopulation (right panel).
(K) Schemes summarizing major fibroblast groups at E12.5 and their location.
